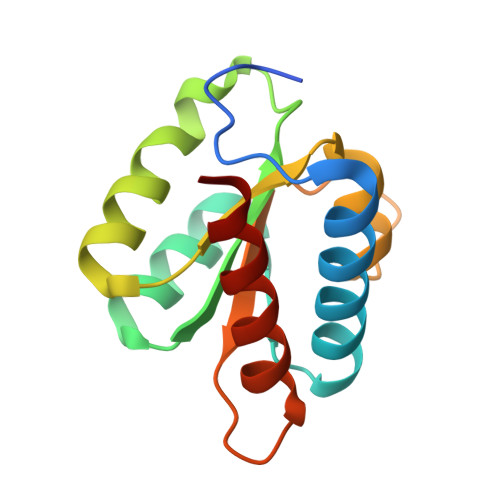
Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control
Bizarro, J., Charron, C., Boulon, S., Westman, B., Pradet-Balade, B., Vandermoere, F., Chagot, M.E., Hallais, M., Ahmad, Y., Leonhardt, H., Lamond, A., Manival, X., Branlant, C., Charpentier, B., Verheggen, C., Bertrand, E.(2014) J Cell Biol 207: 463-480
- PubMed: 25404746
- DOI: https://doi.org/10.1083/jcb.201404160
- Primary Citation of Related Structures:
4NUT - PubMed Abstract:
In vitro, assembly of box C/D small nucleolar ribonucleoproteins (snoRNPs) involves the sequential recruitment of core proteins to snoRNAs. In vivo, however, assembly factors are required (NUFIP, BCD1, and the HSP90-R2TP complex), and it is unknown whether a similar sequential scheme applies. In this paper, we describe systematic quantitative stable isotope labeling by amino acids in cell culture proteomic experiments and the crystal structure of the core protein Snu13p/15.5K bound to a fragment of the assembly factor Rsa1p/NUFIP. This revealed several unexpected features: (a) the existence of a protein-only pre-snoRNP complex containing five assembly factors and two core proteins, 15.5K and Nop58; (b) the characterization of ZNHIT3, which is present in the protein-only complex but gets released upon binding to C/D snoRNAs; (c) the dynamics of the R2TP complex, which appears to load/unload RuvBL AAA(+) adenosine triphosphatase from pre-snoRNPs; and (d) a potential mechanism for preventing premature activation of snoRNP catalytic activity. These data provide a framework for understanding the assembly of box C/D snoRNPs.
- Equipe labellisée Ligue contre le Cancer, Centre National de la Recherche Scientifique Unité Mixte de Recherche 5535, Institut de Génétique Moléculaire de Montpellier, 34293 Montpellier, Cedex 5, France.
Organizational Affiliation: