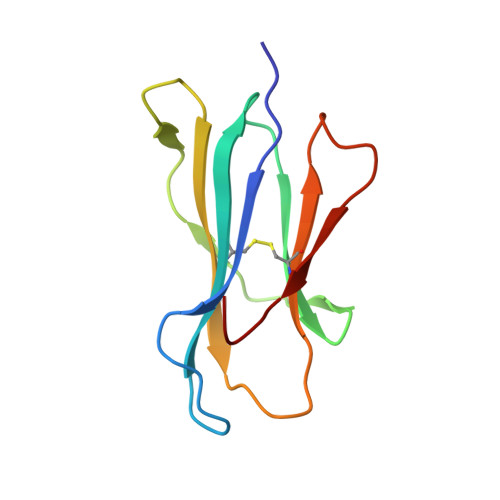
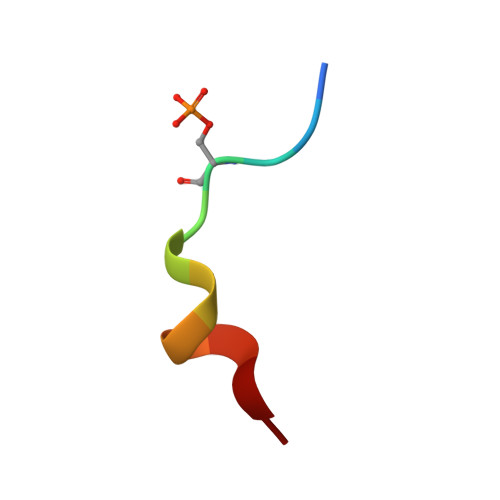
The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status.
Mohammed, F., Stones, D.H., Zarling, A.L., Willcox, C.R., Shabanowitz, J., Cummings, K.L., Hunt, D.F., Cobbold, M., Engelhard, V.H., Willcox, B.E.(2017) Oncotarget 8: 54160-54172
- PubMed: 28903331
- DOI: https://doi.org/10.18632/oncotarget.16952
- Primary Citation of Related Structures:
4NNX, 4NNY, 4NO0, 4NO2, 4NO3, 4NO5 - PubMed Abstract:
Dysregulated post-translational modification provides a source of altered self-antigens that can stimulate immune responses in autoimmunity, inflammation, and cancer. In recent years, phosphorylated peptides have emerged as a group of tumour-associated antigens presented by MHC molecules and recognised by T cells, and represent promising candidates for cancer immunotherapy. However, the impact of phosphorylation on the antigenic identity of phosphopeptide epitopes is unclear. Here we examined this by determining structures of MHC-bound phosphopeptides bearing canonical position 4-phosphorylations in the presence and absence of their phosphate moiety, and examining phosphopeptide recognition by the T cell receptor (TCR). Strikingly, two peptides exhibited major conformational changes upon phosphorylation, involving a similar molecular mechanism, which focussed changes on the central peptide region most critical for T cell recognition. In contrast, a third epitope displayed little conformational alteration upon phosphorylation. In addition, binding studies demonstrated TCR interaction with an MHC-bound phosphopeptide was both epitope-specific and absolutely dependent upon phosphorylation status. These results highlight the critical influence of phosphorylation on the antigenic identity of naturally processed class I MHC epitopes. In doing so they provide a molecular framework for understanding phosphopeptide-specific immune responses, and have implications for the development of phosphopeptide antigen-specific cancer immunotherapy approaches.
- Cancer Immunology and Immunotherapy Centre, Institute of Immunology and Immunotherapy, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK.
Organizational Affiliation: