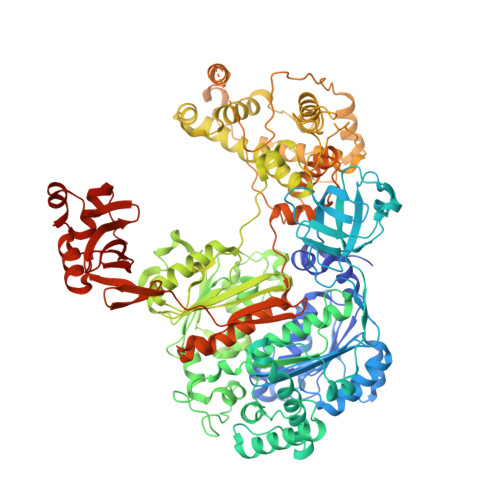
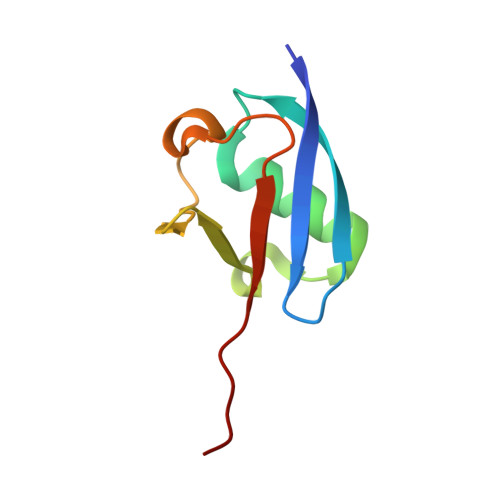
Structure of the ubiquitin-activating enzyme loaded with two ubiquitin molecules.
Schafer, A., Kuhn, M., Schindelin, H.(2014) Acta Crystallogr D Biol Crystallogr 70: 1311-1320
- PubMed: 24816100
- DOI: https://doi.org/10.1107/S1399004714002910
- Primary Citation of Related Structures:
4NNJ - PubMed Abstract:
The activation of ubiquitin by the ubiquitin-activating enzyme Uba1 (E1) constitutes the first step in the covalent modification of target proteins with ubiquitin. This activation is a three-step process in which ubiquitin is adenylated at its C-terminal glycine, followed by the covalent attachment of ubiquitin to a catalytic cysteine residue of Uba1 and the subsequent adenylation of a second ubiquitin. Here, a ubiquitin E1 structure loaded with two ubiquitin molecules is presented for the first time. While one ubiquitin is bound in its adenylated form to the active adenylation domain of E1, the second ubiquitin represents the status after transfer and is covalently linked to the active-site cysteine. The covalently linked ubiquitin enables binding of the E2 enzyme without further modification of the ternary Uba1-ubiquitin2 arrangement. This doubly loaded E1 structure constitutes a missing link in the structural analysis of the ubiquitin-transfer cascade.
- Structural Biology, Rudolf Virchow Center for Experimental Biomedicine, University of Würzburg, Josef-Schneider-Strasse 2, D-97080 Würzburg, Germany.
Organizational Affiliation: