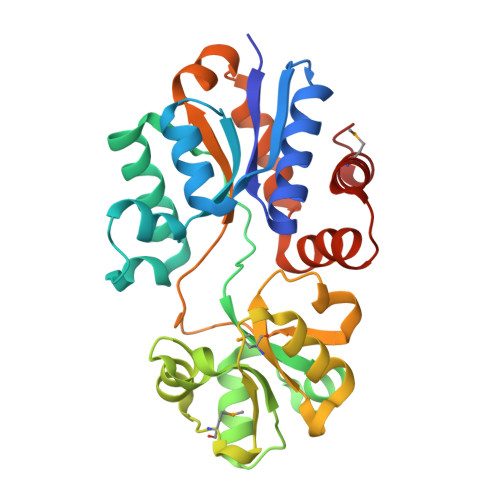
The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation
Nicholls, R., Tkaczuk, K.L., Kagan, O., Chruszcz, M., Domagalski, M.J., Savchenko, A., Joachimiak, A., Murshudov, G., Minor, W., Midwest Center for Structural Genomics (MCSG)To be published.