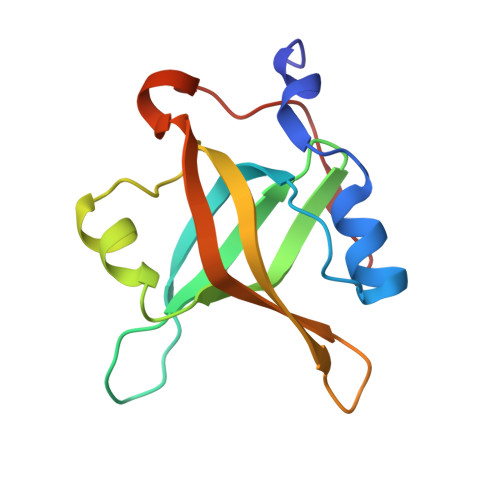
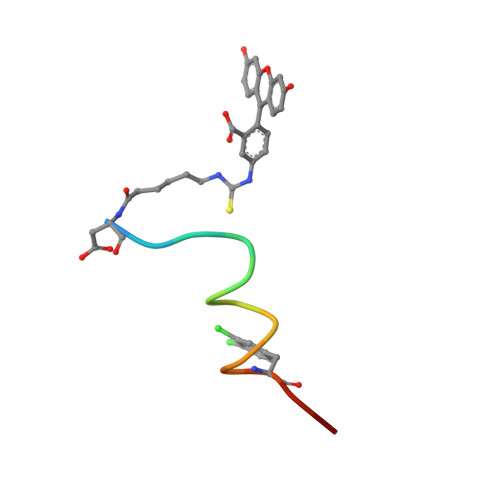
Discovery of a Potent Stapled Helix Peptide That Binds to the 70N Domain of Replication Protein A.
Frank, A.O., Vangamudi, B., Feldkamp, M.D., Souza-Fagundes, E.M., Luzwick, J.W., Cortez, D., Olejniczak, E.T., Waterson, A.G., Rossanese, O.W., Chazin, W.J., Fesik, S.W.(2014) J Med Chem 57: 2455-2461
- PubMed: 24491171
- DOI: https://doi.org/10.1021/jm401730y
- Primary Citation of Related Structures:
4NB3 - PubMed Abstract:
Stapled helix peptides can serve as useful tools for inhibiting protein-protein interactions but can be difficult to optimize for affinity. Here we describe the discovery and optimization of a stapled helix peptide that binds to the N-terminal domain of the 70 kDa subunit of replication protein A (RPA70N). In addition to applying traditional optimization strategies, we employed a novel approach for efficiently designing peptides containing unnatural amino acids. We discovered hot spots in the target protein using a fragment-based screen, identified the amino acid that binds to the hot spot, and selected an unnatural amino acid to incorporate, based on the structure-activity relationships of small molecules that bind to this site. The resulting stapled helix peptide potently and selectively binds to RPA70N, does not disrupt ssDNA binding, and penetrates cells. This peptide may serve as a probe to explore the therapeutic potential of RPA70N inhibition in cancer.
- Department of Biochemistry, Vanderbilt University School of Medicine , Nashville, Tennessee 37232-0146, United States.
Organizational Affiliation: