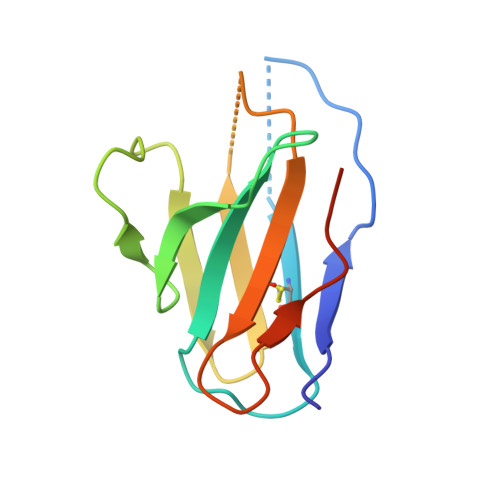
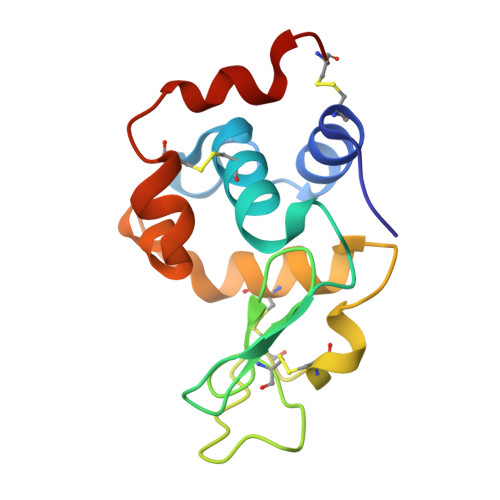
Structural reconstruction of protein ancestry.
Rouet, R., Langley, D.B., Schofield, P., Christie, M., Roome, B., Porebski, B.T., Buckle, A.M., Clifton, B.E., Jackson, C.J., Stock, D., Christ, D.(2017) Proc Natl Acad Sci U S A 114: 3897-3902
- PubMed: 28356519
- DOI: https://doi.org/10.1073/pnas.1613477114
- Primary Citation of Related Structures:
4N1C, 4N1E - PubMed Abstract:
Ancestral protein reconstruction allows the resurrection and characterization of ancient proteins based on computational analyses of sequences of modern-day proteins. Unfortunately, many protein families are highly divergent and not suitable for sequence-based reconstruction approaches. This limitation is exemplified by the antigen receptors of jawed vertebrates (B- and T-cell receptors), heterodimers formed by pairs of Ig domains. These receptors are believed to have evolved from an extinct homodimeric ancestor through a process of gene duplication and diversification; however molecular evidence has so far remained elusive. Here, we use a structural approach and laboratory evolution to reconstruct such molecules and characterize their interaction with antigen. High-resolution crystal structures of reconstructed homodimeric receptors in complex with hen-egg white lysozyme demonstrate how nanomolar affinity binding of asymmetrical antigen is enabled through selective recruitment and structural plasticity within the receptor-binding site. Our results provide structural evidence in support of long-held theories concerning the evolution of antigen receptors, and provide a blueprint for the experimental reconstruction of protein ancestry in the absence of phylogenetic evidence.
- Garvan Institute of Medical Research, Darlinghurst, Sydney, NSW 2010, Australia.
Organizational Affiliation: