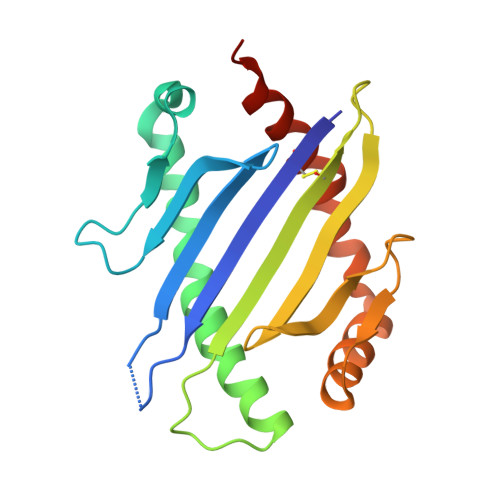
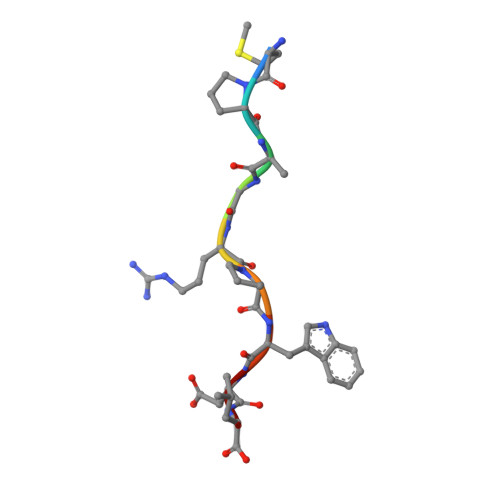
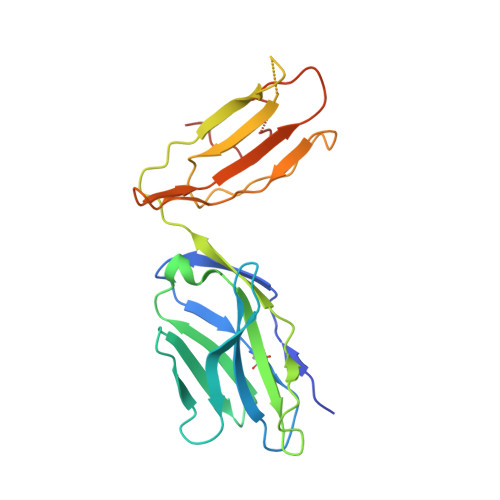
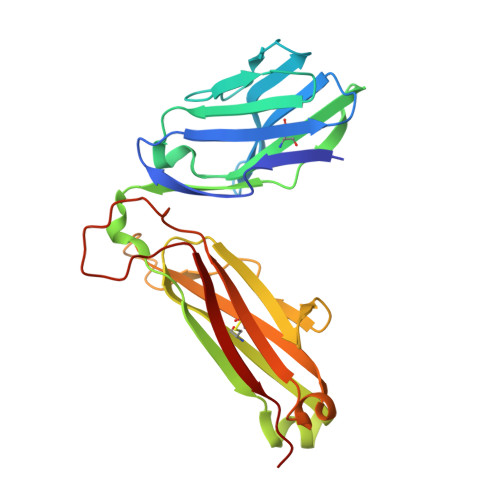
Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Adams, J.J., Narayanan, S., Birnbaum, M.E., Sidhu, S.S., Blevins, S.J., Gee, M.H., Sibener, L.V., Baker, B.M., Kranz, D.M., Garcia, K.C.(2016) Nat Immunol 17: 87-94
- PubMed: 26523866
- DOI: https://doi.org/10.1038/ni.3310
- Primary Citation of Related Structures:
4MS8, 4MVB, 4MXQ, 4N0C, 4N5E - PubMed Abstract:
The T cell antigen receptor (TCR)-peptide-major histocompatibility complex (MHC) interface is composed of conserved and diverse regions, yet the relative contribution of each in shaping recognition by T cells remains unclear. Here we isolated cross-reactive peptides with limited homology, which allowed us to compare the structural properties of nine peptides for a single TCR-MHC pair. The TCR's cross-reactivity was rooted in highly similar recognition of an apical 'hot-spot' position in the peptide with tolerance of sequence variation at ancillary positions. Furthermore, we found a striking structural convergence onto a germline-mediated interaction between the TCR CDR1α region and the MHC α2 helix in twelve TCR-peptide-MHC complexes. Our studies suggest that TCR-MHC germline-mediated constraints, together with a focus on a small peptide hot spot, might place limits on peptide antigen cross-reactivity.
- Howard Hughes Medical Institute, and Departments of Molecular and Cellular Physiology, and Structural Biology, Program in Immunology, Stanford University School of Medicine, Stanford, California, USA.
Organizational Affiliation: