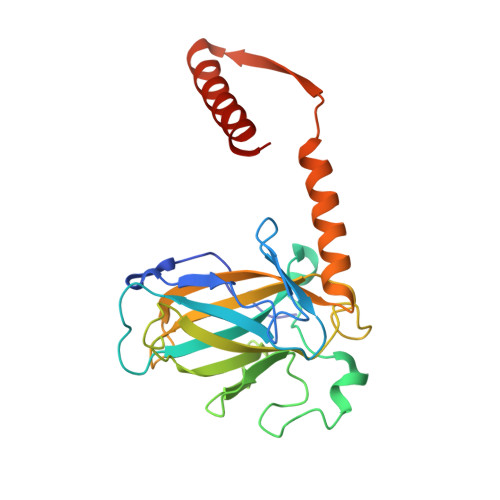
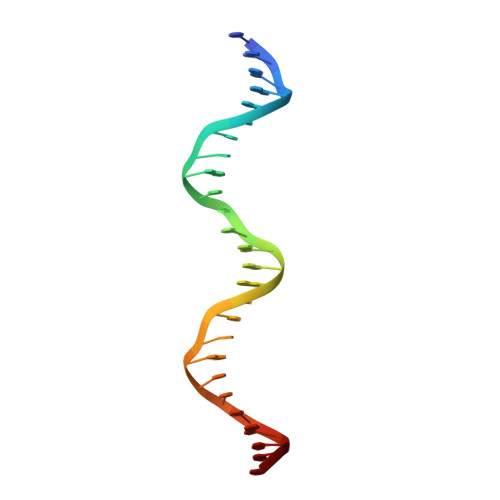
Reversal of the DNA-Binding-Induced Loop L1 Conformational Switch in an Engineered Human p53 Protein.
Emamzadah, S., Tropia, L., Vincenti, I., Falquet, B., Halazonetis, T.D.(2014) J Mol Biology 426: 936-944
- PubMed: 24374182
- DOI: https://doi.org/10.1016/j.jmb.2013.12.020
- Primary Citation of Related Structures:
4MZI, 4MZR - PubMed Abstract:
The gene encoding the p53 tumor suppressor protein, a sequence-specific DNA binding transcription factor, is the most frequently mutated gene in human cancer. Crystal structures of homo-oligomerizing p53 polypeptides with specific DNA suggest that DNA binding is associated with a conformational switch. Specifically, in the absence of DNA, loop L1 of the p53 DNA binding domain adopts an extended conformation, whereas two p53 subunits switch to a recessed loop L1 conformation when bound to DNA as a tetramer. We previously designed a p53 protein, p53FG, with amino substitutions S121F and V122G targeting loop L1. These two substitutions enhanced the affinity of p53 for specific DNA yet, counterintuitively, decreased the residency time of p53 on DNA. Here, we confirmed these DNA binding properties of p53FG using a different method. We also determined by crystallography the structure of p53FG in its free state and bound to DNA as a tetramer. In the free state, loop L1 adopted a recessed conformation, whereas upon DNA binding, two subunits switched to the extended loop L1 conformation, resulting in a final structure that was very similar to that of wild-type p53 bound to DNA. Thus, altering the apo structure of p53 changed its DNA binding properties, even though the DNA-bound structure was not altered.
- Department of Molecular Biology, University of Geneva, 1205 Geneva, Switzerland.
Organizational Affiliation: