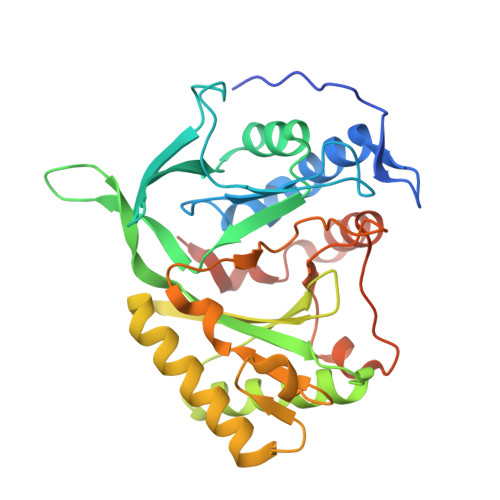
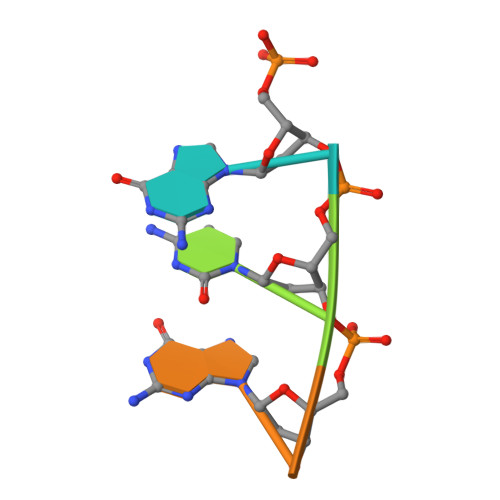
Molecular Basis for DNA Double-Strand Break Annealing and Primer Extension by an NHEJ DNA Polymerase.
Brissett, N.C., Martin, M.J., Bartlett, E.J., Bianchi, J., Blanco, L., Doherty, A.J.(2013) Cell Rep 5: 1108-1120
- PubMed: 24239356
- DOI: https://doi.org/10.1016/j.celrep.2013.10.016
- Primary Citation of Related Structures:
4MKY - PubMed Abstract:
Nonhomologous end-joining (NHEJ) is one of the major DNA double-strand break (DSB) repair pathways. The mechanisms by which breaks are competently brought together and extended during NHEJ is poorly understood. As polymerases extend DNA in a 5'-3' direction by nucleotide addition to a primer, it is unclear how NHEJ polymerases fill in break termini containing 3' overhangs that lack a primer strand. Here, we describe, at the molecular level, how prokaryotic NHEJ polymerases configure a primer-template substrate by annealing the 3' overhanging strands from opposing breaks, forming a gapped intermediate that can be extended in trans. We identify structural elements that facilitate docking of the 3' ends in the active sites of adjacent polymerases and reveal how the termini act as primers for extension of the annealed break, thus explaining how such DSBs are extended in trans. This study clarifies how polymerases couple break-synapsis to catalysis, providing a molecular mechanism to explain how primer extension is achieved on DNA breaks.
- Genome Damage and Stability Centre, University of Sussex, Brighton BN1 9RQ, UK.
Organizational Affiliation: