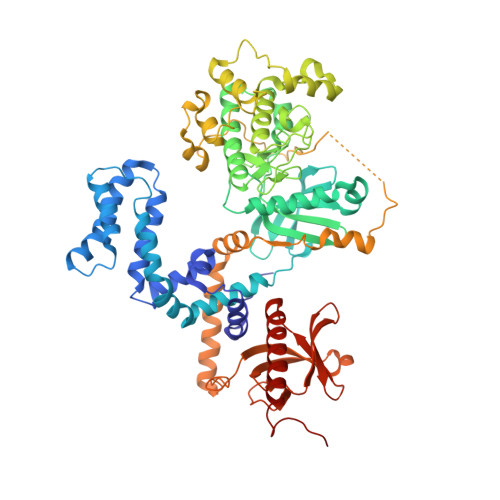
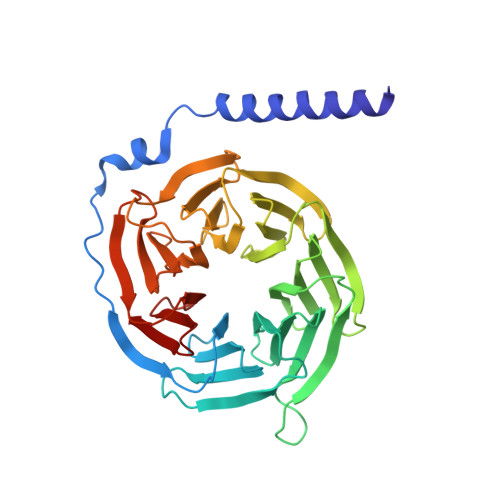
Structural and functional analysis of g protein-coupled receptor kinase inhibition by paroxetine and a rationally designed analog.
Homan, K.T., Wu, E., Wilson, M.W., Singh, P., Larsen, S.D., Tesmer, J.J.(2014) Mol Pharmacol 85: 237-248
- PubMed: 24220010
- DOI: https://doi.org/10.1124/mol.113.089631
- Primary Citation of Related Structures:
4L9I, 4MK0 - PubMed Abstract:
Recently we identified the serotonin reuptake inhibitor paroxetine as an inhibitor of G protein-coupled receptor kinase 2 (GRK2) that improves cardiac performance in live animals. Paroxetine exhibits up to 50-fold selectivity for GRK2 versus other GRKs. A better understanding of the molecular basis of this selectivity is important for the development of even more selective and potent small molecule therapeutics and chemical genetic probes. We first sought to understand the molecular mechanisms underlying paroxetine selectivity among GRKs. We directly measured the K(D) for paroxetine and assessed its mechanism of inhibition for each of the GRK subfamilies and then determined the atomic structure of its complex with GRK1, the most weakly inhibited GRK tested. Our results suggest that the selectivity of paroxetine for GRK2 largely reflects its lower affinity for adenine nucleotides. Thus, stabilization of off-pathway conformational states unique to GRK2 will likely be key for the development of even more selective inhibitors. Next, we designed a benzolactam derivative of paroxetine that has optimized interactions with the hinge of the GRK2 kinase domain. The crystal structure of this compound in complex with GRK2 confirmed the predicted interactions. Although the benzolactam derivative did not significantly alter potency of inhibition among GRKs, it exhibited 20-fold lower inhibition of serotonin reuptake. However, there was an associated increase in the potency for inhibition of other AGC kinases, suggesting that the unconventional hydrogen bond formed by the benzodioxole ring of paroxetine is better accommodated by GRKs.
- Life Sciences Institute and the Departments of Pharmacology and Biological Sciences (K.T.H., E.W., P.S., J.J.G.T.), and Vahlteich Medicinal Chemistry Core and the Department of Medicinal Chemistry, University of Michigan, Ann Arbor, Michigan (M.W.W., S.D.L.).
Organizational Affiliation: