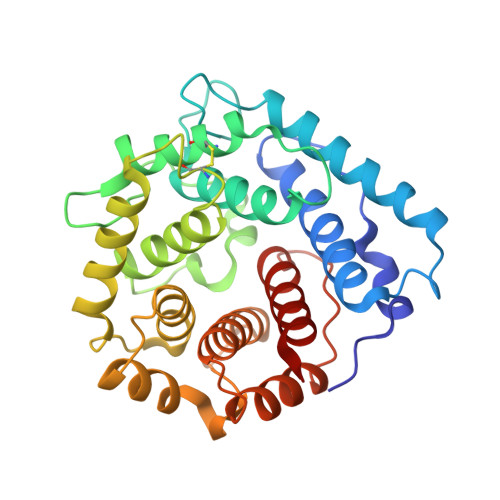
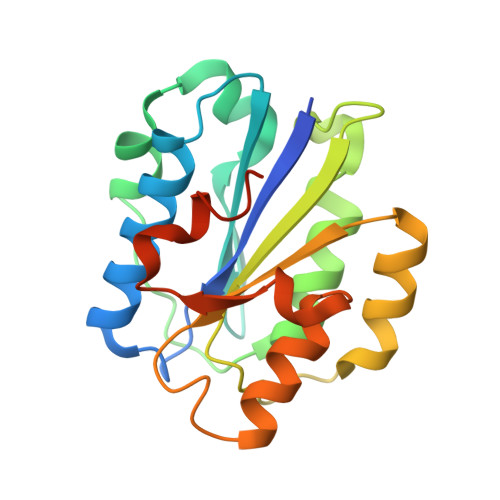
Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3.
Bajic, G., Yatime, L., Sim, R.B., Vorup-Jensen, T., Andersen, G.R.(2013) Proc Natl Acad Sci U S A 110: 16426-16431
- PubMed: 24065820
- DOI: https://doi.org/10.1073/pnas.1311261110
- Primary Citation of Related Structures:
4M76 - PubMed Abstract:
Complement receptors (CRs), expressed notably on myeloid and lymphoid cells, play an essential function in the elimination of complement-opsonized pathogens and apoptotic/necrotic cells. In addition, these receptors are crucial for the cross-talk between the innate and adaptive branches of the immune system. CR3 (also known as Mac-1, integrin αMβ2, or CD11b/CD18) is expressed on all macrophages and recognizes iC3b on complement-opsonized objects, enabling their phagocytosis. We demonstrate that the C3d moiety of iC3b harbors the binding site for the CR3 αI domain, and our structure of the C3d:αI domain complex rationalizes the CR3 selectivity for iC3b. Based on extensive structural analysis, we suggest that the choice between a ligand glutamate or aspartate for coordination of a receptor metal ion-dependent adhesion site-bound metal ion is governed by the secondary structure of the ligand. Comparison of our structure to the CR2:C3d complex and the in vitro formation of a stable CR3:C3d:CR2 complex suggests a molecular mechanism for the hand-over of CR3-bound immune complexes from macrophages to CR2-presenting cells in lymph nodes.
- Departments of Molecular Biology and Genetics and Biomedicine, Aarhus University, DK-8000 Aarhus, Denmark.
Organizational Affiliation: