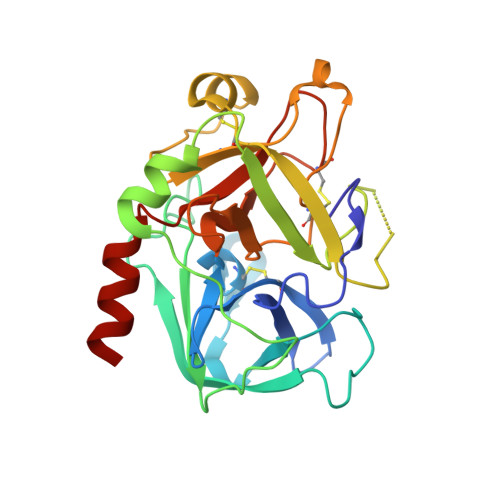
5-Chlorothiophene-2-carboxylic acid [(S)-2-[2-methyl-3-(2-oxopyrrolidin-1-yl)benzenesulfonylamino]-3-(4-methylpiperazin-1-yl)-3-oxopropyl]amide (SAR107375), a selective and potent orally active dual thrombin and factor Xa inhibitor.
Meneyrol, J., Follmann, M., Lassalle, G., Wehner, V., Barre, G., Rousseaux, T., Altenburger, J.M., Petit, F., Bocskei, Z., Schreuder, H., Alet, N., Herault, J.P., Millet, L., Dol, F., Florian, P., Schaeffer, P., Sadoun, F., Klieber, S., Briot, C., Bono, F., Herbert, J.M.(2013) J Med Chem 56: 9441-9456
- PubMed: 24175584
- DOI: https://doi.org/10.1021/jm4005835
- Primary Citation of Related Structures:
4BTI, 4BTT, 4BTU, 4LOY, 4LXB - PubMed Abstract:
Compound 15 (SAR107375), a novel potent dual thrombin and factor Xa inhibitor resulted from a rational optimization process. Starting from compound 14, with low factor Xa and modest anti-thrombin inhibitory activities (IC50's of 3.5 and 0.39 μM, respectively), both activities were considerably improved, notably through the incorporation of a neutral chlorothiophene P1 fragment and tuning of P2 and P3-P4 fragments. Final optimization of metabolic stability with microsomes led to the identification of 15, which displays strong activity in vitro vs factor Xa and thrombin (with Ki's of 1 and 8 nM, respectively). In addition 15 presents good selectivity versus related serine proteases (roughly 300-fold), including trypsin (1000-fold), and is very active (0.39 μM) in the thrombin generation time (TGT) coagulation assay in human platelet rich plasma (PRP). Potent in vivo activity in a rat model of venous thrombosis following iv and, more importantly, po administration was also observed (ED50 of 0.07 and 2.8 mg/kg, respectively). Bleeding liability was reduced in the rat wire coil model, more relevant to arterial thrombosis, with 15 (blood loss increase of 2-fold relative to the ED80 value) compared to rivaroxaban 2 and dabigatran etexilate 1a.
- Sanofi-Aventis R&D , 195 Route d'Espagne, 31036 Toulouse Cedex, France.
Organizational Affiliation: