Structural Insights on the Bacteriolytic and Self-protection Mechanism of Muramidase Effector Tse3 in Pseudomonas aeruginosa
Li, L., Zhang, W., Liu, Q., Gao, Y., Gao, Y., Wang, Y., Wang, D.Z., Li, Z., Wang, T.(2013) J Biological Chem 288: 30607-30613
- PubMed: 24025333
- DOI: https://doi.org/10.1074/jbc.C113.506097
- Primary Citation of Related Structures:
4LUQ - PubMed Abstract:
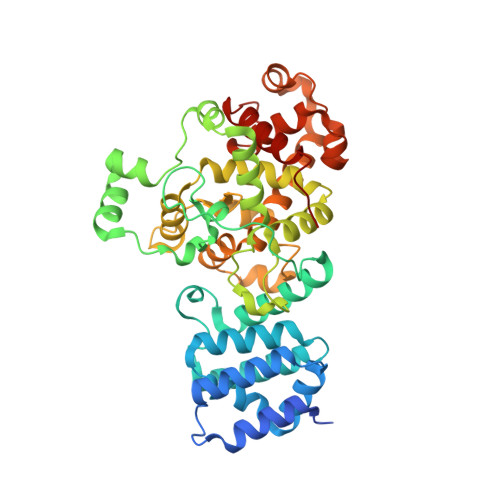
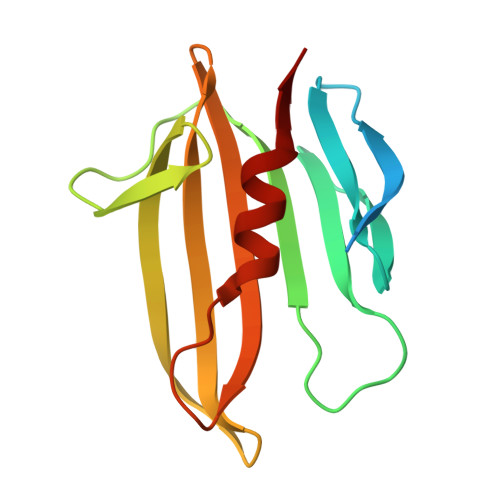
The warfare among microbial species as well as between pathogens and hosts is fierce, complicated, and continuous. In Pseudomonas aeruginosa, the muramidase effector Tse3 (Type VI secretion exported 3) can be injected into the periplasm of neighboring bacterial competitors by a Type VI secretion apparatus, eventually leading to cell lysis and death. However, P. aeruginosa protects itself from lysis by expressing immune protein Tsi3 (Type six secretion immunity 3). Here, we report the crystal structure of the Tse3-Tsi3 complex at 1.8 Å resolution, revealing that Tse3 possesses one open accessible, goose-type lysozyme-like domain with peptidoglycan hydrolysis activity. Calcium ions bind specifically in the Tse3 active site and are identified to be crucial for its bacteriolytic activity. In combination with biochemical studies, the structural basis of self-protection mechanism of Tsi3 is also elucidated, thus providing an understanding and new insights into the effectors of Type VI secretion system.
- From the Laboratory for Computational Chemistry and Drug Design and.
Organizational Affiliation: