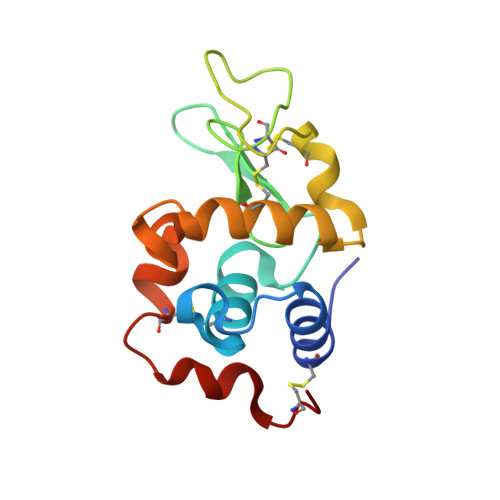
Carboplatin binding to a model protein in non-NaCl conditions to eliminate partial conversion to cisplatin, and the use of different criteria to choose the resolution limit
Tanley, S.W.M., Diederichs, K., Kroon-Batenburg, L.M.J., Schreurs, A.M.M., Helliwell, J.R.To be published.