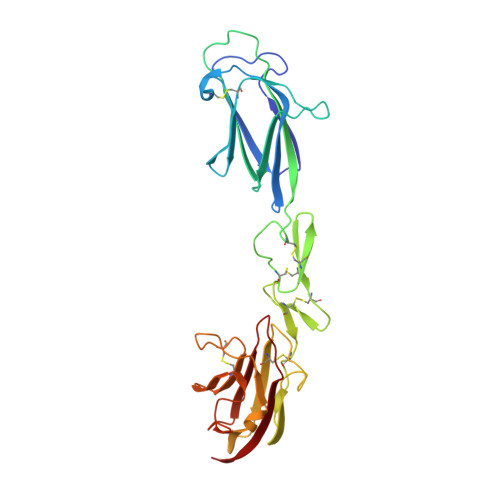
Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Venkatraman Girija, U., Gingras, A.R., Marshall, J.E., Panchal, R., Sheikh, M.A., Gal, P., Schwaeble, W.J., Mitchell, D.A., Moody, P.C., Wallis, R.(2013) Proc Natl Acad Sci U S A 110: 13916-13920
- PubMed: 23922389
- DOI: https://doi.org/10.1073/pnas.1311113110
- Primary Citation of Related Structures:
4LMF, 4LOR, 4LOS, 4LOT - PubMed Abstract:
Complement component C1, the complex that initiates the classical pathway of complement activation, is a 790-kDa assembly formed from the target-recognition subcomponent C1q and the modular proteases C1r and C1s. The proteases are elongated tetramers that become more compact when they bind to the collagen-like domains of C1q. Here, we describe a series of structures that reveal how the subcomponents associate to form C1. A complex between C1s and a collagen-like peptide containing the C1r/C1s-binding motif of C1q shows that the collagen binds to a shallow groove via a critical lysine side chain that contacts Ca(2+)-coordinating residues. The data explain the Ca(2+)-dependent binding mechanism, which is conserved in C1r and also in mannan-binding lectin-associated serine proteases, the serine proteases of the lectin pathway activation complexes. In an accompanying structure, C1s forms a compact ring-shaped tetramer featuring a unique head-to-tail interaction at its center that replicates the likely arrangement of C1r/C1s polypeptides in the C1 complex. Additional structures reveal how C1s polypeptides are positioned to enable activation by C1r and interaction with the substrate C4 inside the cage-like assembly formed by the collagenous stems of C1q. Together with previously determined structures of C1r fragments, the results reported here provide a structural basis for understanding the early steps of complement activation via the classical pathway.
- Department of Infection, University of Leicester, Leicester LE1 9HN, United Kingdom.
Organizational Affiliation: