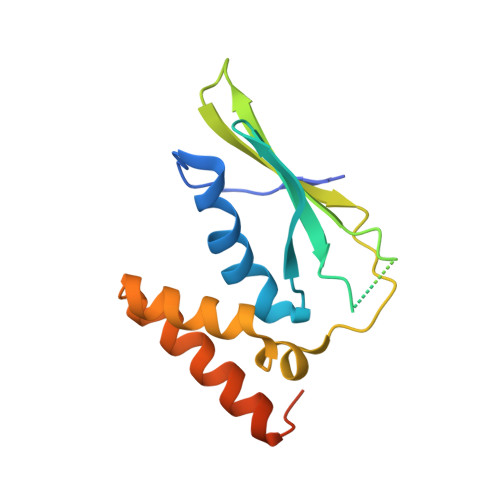
Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2.
Poor, C.B., Wegner, S.V., Li, H., Dlouhy, A.C., Schuermann, J.P., Sanishvili, R., Hinshaw, J.R., Riggs-Gelasco, P.J., Outten, C.E., He, C.(2014) Proc Natl Acad Sci U S A 111: 4043-4048
- PubMed: 24591629
- DOI: https://doi.org/10.1073/pnas.1318869111
- Primary Citation of Related Structures:
4LMG - PubMed Abstract:
The paralogous iron-responsive transcription factors Aft1 and Aft2 (activators of ferrous transport) regulate iron homeostasis in Saccharomyces cerevisiae by activating expression of iron-uptake and -transport genes when intracellular iron is low. We present the previously unidentified crystal structure of Aft2 bound to DNA that reveals the mechanism of DNA recognition via specific interactions of the iron-responsive element with a Zn(2+)-containing WRKY-GCM1 domain in Aft2. We also show that two Aft2 monomers bind a [2Fe-2S] cluster (or Fe(2+)) through a Cys-Asp-Cys motif, leading to dimerization of Aft2 and decreased DNA-binding affinity. Furthermore, we demonstrate that the [2Fe-2S]-bridged heterodimer formed between glutaredoxin-3 and the BolA-like protein Fe repressor of activation-2 transfers a [2Fe-2S] cluster to Aft2 that facilitates Aft2 dimerization. Previous in vivo findings strongly support the [2Fe-2S] cluster-induced dimerization model; however, given the available evidence, Fe(2+)-induced Aft2 dimerization cannot be completely ruled out as an alternative Aft2 inhibition mechanism. Taken together, these data provide insight into the molecular mechanism for iron-dependent transcriptional regulation of Aft2 and highlight the key role of Fe-S clusters as cellular iron signals.
- Department of Chemistry and Institute for Biophysical Dynamics, The University of Chicago, Chicago, IL 60637.
Organizational Affiliation: