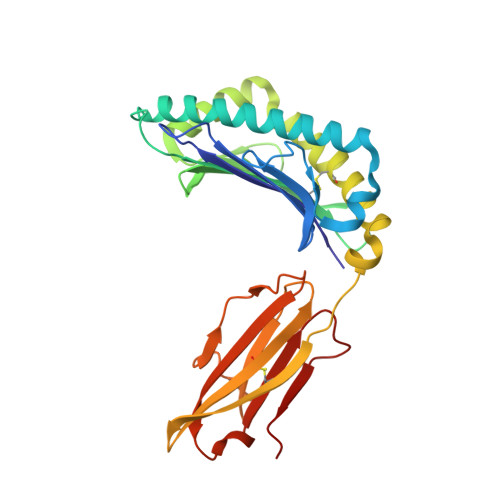
Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
Halabelian, L., Ricagno, S., Giorgetti, S., Santambrogio, C., Barbiroli, A., Pellegrino, S., Achour, A., Grandori, R., Marchese, L., Raimondi, S., Mangione, P.P., Esposito, G., Al-Shawi, R., Simons, J.P., Speck, I., Stoppini, M., Bolognesi, M., Bellotti, V.(2014) J Biological Chem 289: 3318-3327
- PubMed: 24338476
- DOI: https://doi.org/10.1074/jbc.M113.524157
- Primary Citation of Related Structures:
4L29, 4L3C - PubMed Abstract:
To form extracellular aggregates, amyloidogenic proteins bypass the intracellular quality control, which normally targets unfolded/aggregated polypeptides. Human D76N β2-microglobulin (β2m) variant is the prototype of unstable and amyloidogenic protein that forms abundant extracellular fibrillar deposits. Here we focus on the role of the class I major histocompatibility complex (MHCI) in the intracellular stabilization of D76N β2m. Using biophysical and structural approaches, we show that the MHCI containing D76N β2m (MHCI76) displays stability, dissociation patterns, and crystal structure comparable with those of the MHCI with wild type β2m. Conversely, limited proteolysis experiments show a reduced protease susceptibility for D76N β2m within the MHCI76 as compared with the free variant, suggesting that the MHCI has a chaperone-like activity in preventing D76N β2m degradation within the cell. Accordingly, D76N β2m is normally assembled in the MHCI and circulates as free plasma species in a transgenic mouse model.
- From the Dipartimento di Bioscienze, Università di Milano, Via Celoria 26, 20133 Milano, Italy.
Organizational Affiliation: