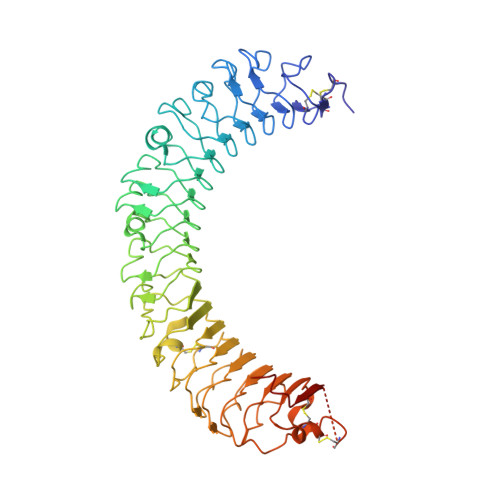
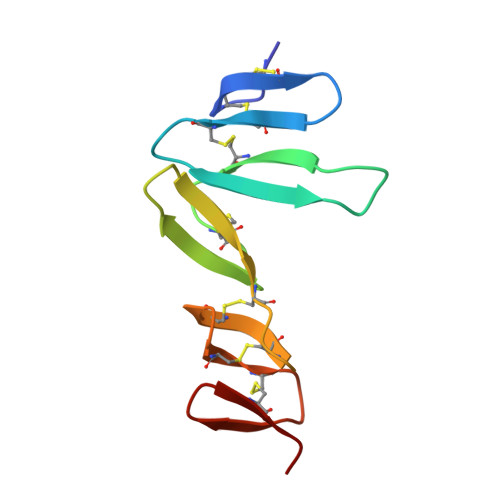
Structural basis for R-spondin recognition by LGR4/5/6 receptors
Wang, D.L., Huang, B., Zhang, S., Yu, X., Wu, W., Wang, X.Q.(2013) Genes Dev 27: 1339-1344
- PubMed: 23756652
- DOI: https://doi.org/10.1101/gad.219360.113
- Primary Citation of Related Structures:
4KT1 - PubMed Abstract:
The R-spondin (RSPO) family of secreted proteins (RSPO1-RSPO4) has pleiotropic functions in development and stem cell growth by strongly enhancing Wnt pathway activation. Recently, leucine-rich repeat-containing G-protein-coupled receptor 4 (LGR4), LGR5, and LGR6 have been identified as receptors for RSPOs. Here we report the complex structure of the LGR4 extracellular domain (ECD) with the RSPO1 N-terminal fragment (RSPO1-2F) containing two adjacent furin-like cysteine-rich domains (FU-CRDs). The LGR4-ECD adopts the anticipated TLR horseshoe structure and uses its concave surface close to the N termini to bind RSPO1-2F. Both the FU-CRD1 and FU-CRD2 domains of RSPO1 contribute to LGR4 interaction, and binding and cellular assays identified critical RSPO1 residues for its biological activities. Our results define the molecular mechanism by which the LGR4/5/6 receptors recognize RSPOs and also provide structural insights into the signaling difference between the LGR4/5/6 receptors and other members in the LGR family.
- Ministry of Education Key Laboratory of Protein Science, Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, PR China.
Organizational Affiliation: