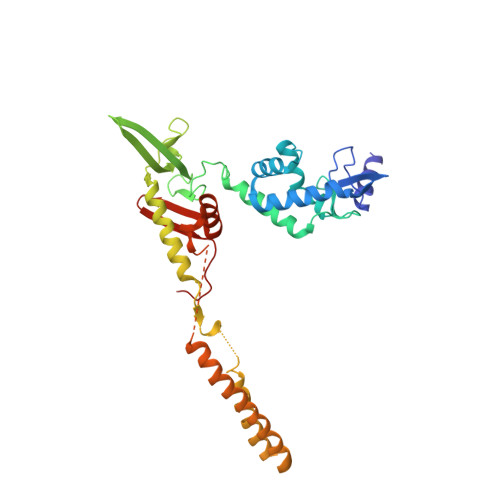
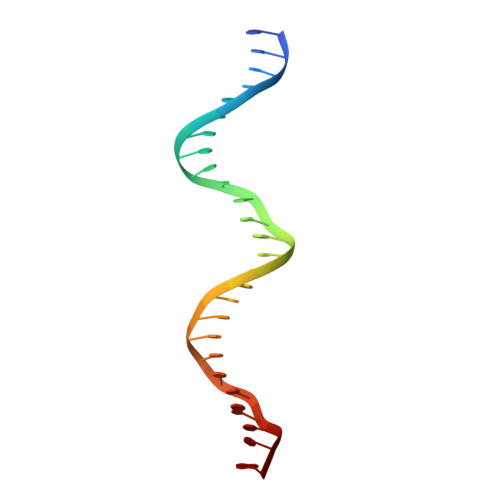
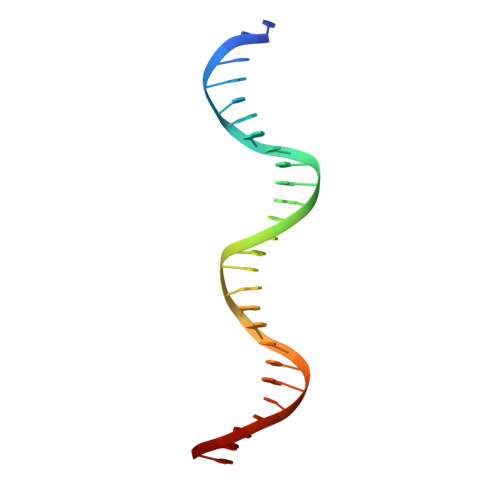
Attachment site recognition and regulation of directionality by the serine integrases.
Rutherford, K., Yuan, P., Perry, K., Sharp, R., Van Duyne, G.D.(2013) Nucleic Acids Res 41: 8341-8356
- PubMed: 23821671
- DOI: https://doi.org/10.1093/nar/gkt580
- Primary Citation of Related Structures:
4KIS - PubMed Abstract:
Serine integrases catalyze the integration of bacteriophage DNA into a host genome by site-specific recombination between 'attachment sites' in the phage (attP) and the host (attB). The reaction is highly directional; the reverse excision reaction between the product attL and attR sites does not occur in the absence of a phage-encoded factor, nor does recombination occur between other pairings of attachment sites. A mechanistic understanding of how these enzymes achieve site-selectivity and directionality has been limited by a lack of structural models. Here, we report the structure of the C-terminal domains of a serine integrase bound to an attP DNA half-site. The structure leads directly to models for understanding how the integrase-bound attP and attB sites differ, why these enzymes preferentially form attP × attB synaptic complexes to initiate recombination, and how attL × attR recombination is prevented. In these models, different domain organizations on attP vs. attB half-sites allow attachment-site specific interactions to form between integrase subunits via an unusual protruding coiled-coil motif. These interactions are used to preferentially synapse integrase-bound attP and attB and inhibit synapsis of integrase-bound attL and attR. The results provide a structural framework for understanding, testing and engineering serine integrase function.
- Department of Biochemistry and Biophysics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA and NE-CAT and Department of Chemistry and Chemical Biology, Cornell University, Building 436E, Argonne National Laboratory, 9700 S. Cass Avenue, Argonne, IL 60439, USA.
Organizational Affiliation: