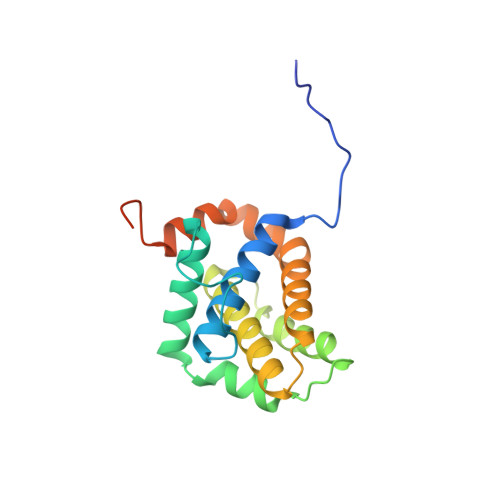
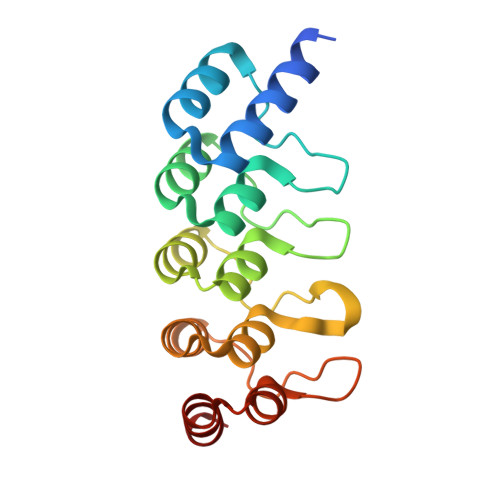
Co-Crystallization with Conformation-Specific Designed Ankyrin Repeat Proteins Explains the Conformational Flexibility of BCL-W
Schilling, J., Schoppe, J., Sauer, E., Pluckthun, A.(2014) J Mol Biology 426: 2346-2362
- PubMed: 24747052
- DOI: https://doi.org/10.1016/j.jmb.2014.04.010
- Primary Citation of Related Structures:
4K5A, 4K5B - PubMed Abstract:
BCL-W is a member of the BCL-2 family of anti-apoptotic proteins. A key event in the regulation of apoptosis is the heterodimerization between anti-apoptotic and pro-apoptotic family members, which involves a conserved surface-exposed groove on the anti-apoptotic proteins. Crystal structures of the ligand binding-competent conformation exist for all anti-apoptotic family members, with the exception of BCL-W, due to the flexibility of the BCL-W groove region. Existing structures had suggested major deviations of the BCL-W groove region from the otherwise structurally highly related remaining anti-apoptotic family members. To capture its ligand binding-competent conformation by counteracting the conformational flexibility of the BCL-W groove, we had selected high-affinity groove-binding designed ankyrin repeat proteins (DARPins) using ribosome display. We now determined two high-resolution crystal structures of human BCL-W in complex with different DARPins at resolutions 1.5 and 1.85Å, in which the structure of BCL-W is virtually identical, and BCL-W adopts a conformation extremely similar to the ligand-free conformation of its closest relative BCL-XL in both structures. However, distinct differences to all previous BCL-W structures are evident, notably in the ligand-binding region. We provide the first structural explanation for the conformational flexibility of the BCL-W groove region in comparison to other BCL-2 family members. Due to the importance of the anti-apoptotic BCL-2 family as drug targets, the presented crystal structure of ligand binding-competent BCL-W may serve as a valuable basis for structure-based drug design in the future and provides a missing piece for the structural characterization of this protein family.
- Biochemisches Institut, Universität Zürich, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland.
Organizational Affiliation: