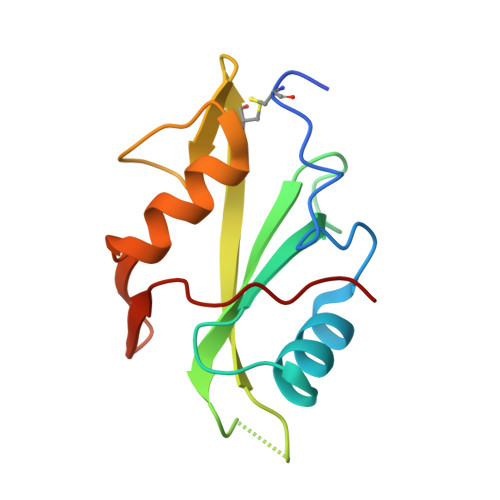
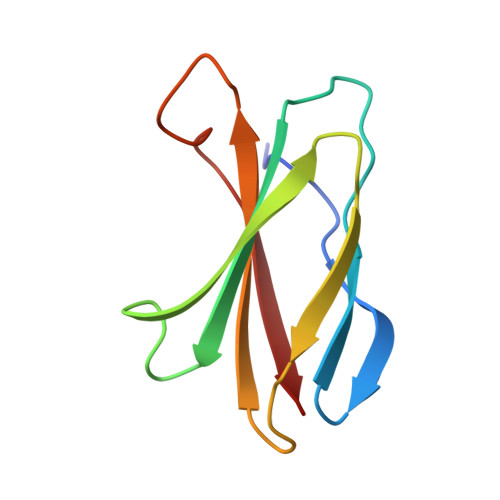
Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains.
Sha, F., Gencer, E.B., Georgeon, S., Koide, A., Yasui, N., Koide, S., Hantschel, O.(2013) Proc Natl Acad Sci U S A 110: 14924-14929
- PubMed: 23980151
- DOI: https://doi.org/10.1073/pnas.1303640110
- Primary Citation of Related Structures:
4JE4, 4JEG - PubMed Abstract:
The dysregulated tyrosine kinase BCR-ABL causes chronic myelogenous leukemia in humans and forms a large multiprotein complex that includes the Src-homology 2 (SH2) domain-containing phosphatase 2 (SHP2). The expression of SHP2 is necessary for BCR-ABL-dependent oncogenic transformation, but the precise signaling mechanisms of SHP2 are not well understood. We have developed binding proteins, termed monobodies, for the N- and C-terminal SH2 domains of SHP2. Intracellular expression followed by interactome analysis showed that the monobodies are essentially monospecific to SHP2. Two crystal structures revealed that the monobodies occupy the phosphopeptide-binding sites of the SH2 domains and thus can serve as competitors of SH2-phosphotyrosine interactions. Surprisingly, the segments of both monobodies that bind to the peptide-binding grooves run in the opposite direction to that of canonical phosphotyrosine peptides, which may contribute to their exquisite specificity. When expressed in cells, monobodies targeting the N-SH2 domain disrupted the interaction of SHP2 with its upstream activator, the Grb2-associated binder 2 adaptor protein, suggesting decoupling of SHP2 from the BCR-ABL protein complex. Inhibition of either N-SH2 or C-SH2 was sufficient to inhibit two tyrosine phosphorylation events that are critical for SHP2 catalytic activity and to block ERK activation. In contrast, targeting the N-SH2 or C-SH2 revealed distinct roles of the two SH2 domains in downstream signaling, such as the phosphorylation of paxillin and signal transducer and activator of transcription 5. Our results delineate a hierarchy of function for the SH2 domains of SHP2 and validate monobodies as potent and specific antagonists of protein-protein interactions in cancer cells.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637 and Swiss Institute for Experimental Cancer Research, School of Life Sciences, École Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland.
Organizational Affiliation: