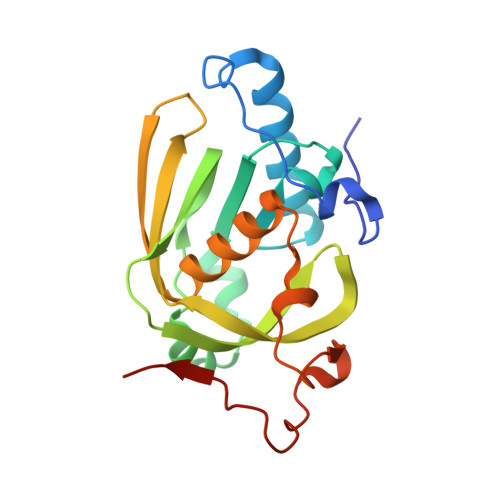
Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Fieulaine, S., Desmadril, M., Meinnel, T., Giglione, C.(2014) Acta Crystallogr D Biol Crystallogr 70: 242-252
- PubMed: 24531459
- DOI: https://doi.org/10.1107/S1399004713026461
- Primary Citation of Related Structures:
4JE6, 4JE7, 4JE8 - PubMed Abstract:
Peptide deformylases (PDFs), which are essential and ubiquitous enzymes involved in the removal of the N-formyl group from nascent chains, are classified into four subtypes based on the structural and sequence similarity of specific conserved domains. All PDFs share a similar three-dimensional structure, are functionally interchangeable in vivo and display similar properties in vitro, indicating that their molecular mechanism has been conserved during evolution. The human mitochondrial PDF is the only exception as despite its conserved fold it reveals a unique substrate-binding pocket together with an unusual kinetic behaviour. Unlike human PDF, the closely related mitochondrial PDF1As from plants have catalytic efficiencies and enzymatic parameters that are similar to those of other classes of PDFs. Here, the aim was to identify the structural basis underlying the properties of human PDF compared with all other PDFs by focusing on plant mitochondrial PDF1A. The construction of a chimaera composed of plant PDF1A with the nonrandom substitutions found in a conserved motif of its human homologue converted it into an enzyme with properties similar to the human enzyme, indicating the crucial role of these positions. The crystal structure of this human-like plant PDF revealed that substitution of two residues leads to a reduction in the volume of the ligand-binding site together with the introduction of negative charges, unravelling the origin of the weak affinity of human PDF for its substrate. In addition, the substitution of the two residues of human PDF modifies the transition state of the reaction through alteration of the network of interactions between the catalytic residues and the substrate, leading to an overall reduced reaction rate.
- CNRS, Centre de Recherche de Gif, Institut des Sciences du Végétal, Bâtiment 23A, 1 Avenue de la Terrasse, 91198 Gif-sur-Yvette CEDEX, France.
Organizational Affiliation: