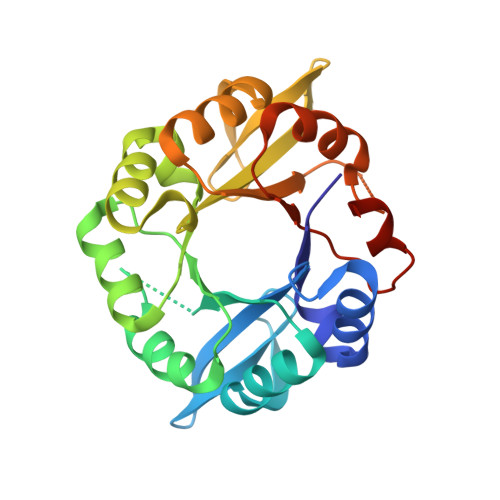
Establishing catalytic activity on an artificial ( beta alpha )8-barrel protein designed from identical half-barrels.
Sperl, J.M., Rohweder, B., Rajendran, C., Sterner, R.(2013) FEBS Lett 587: 2798-2805
- PubMed: 23806364
- DOI: https://doi.org/10.1016/j.febslet.2013.06.022
- Primary Citation of Related Structures:
4J9J - PubMed Abstract:
It has been postulated that the ubiquitous (βα)8-barrel enzyme fold has evolved by duplication and fusion of an ancestral (βα)4-half-barrel. We have previously reconstructed this process in the laboratory by fusing two copies of the C-terminal half-barrel HisF-C of imidazole glycerol phosphate synthase (HisF). The resulting construct HisF-CC was stepwise stabilized to Sym1 and Sym2, which are extremely robust but catalytically inert proteins. Here, we report on the generation of a circular permutant of Sym2 and the establishment of a sugar isomerization reaction on its scaffold. Our results demonstrate that duplication and mutagenesis of (βα)4-half-barrels can readily lead to a stable and catalytically active (βα)8-barrel enzyme.
- Institute of Biophysics and Physical Biochemistry, University of Regensburg, Universitätsstrasse 31, D-93053 Regensburg, Germany.
Organizational Affiliation: