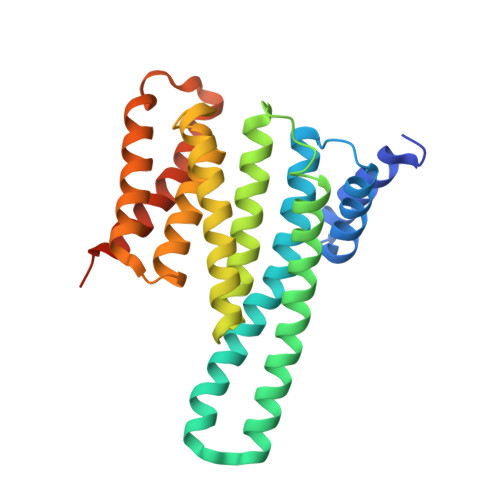
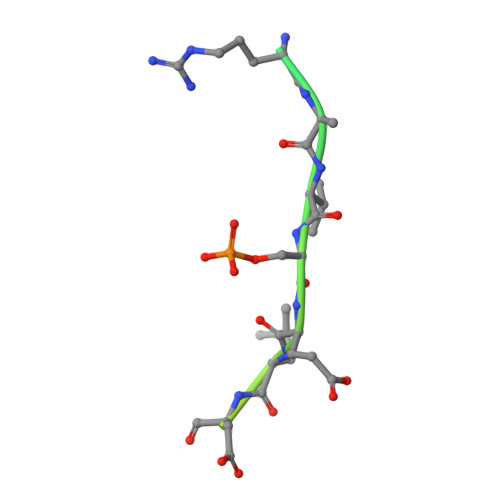
The N-terminal sequence of tyrosine hydroxylase is a conformationally versatile motif that binds 14-3-3 proteins and membranes.
Skjevik, A.A., Mileni, M., Baumann, A., Halskau, O., Teigen, K., Stevens, R.C., Martinez, A.(2014) J Mol Biology 426: 150-168
- PubMed: 24055376
- DOI: https://doi.org/10.1016/j.jmb.2013.09.012
- Primary Citation of Related Structures:
4J6S - PubMed Abstract:
Tyrosine hydroxylase (TH) catalyzes the rate-limiting step in the synthesis of catecholamine neurotransmitters, and a reduction in TH activity is associated with several neurological diseases. Human TH is regulated, among other mechanisms, by Ser19-phosphorylation-dependent interaction with 14-3-3 proteins. The N-terminal sequence (residues 1-43), which corresponds to an extension to the TH regulatory domain, also interacts with negatively charged membranes. By using X-ray crystallography together with molecular dynamics simulations and structural bioinformatics analysis, we have probed the conformations of the Ser19-phosphorylated N-terminal peptide [THp-(1-43)] bound to 14-3-3γ, free in solution and bound to a phospholipid bilayer, and of the unphosphorylated peptide TH-(1-43) both free and bilayer bound. As seen in the crystal structure of THp-(1-43) complexed with 14-3-3γ, the region surrounding pSer19 adopts an extended conformation in the bound state, whereas THp-(1-43) adopts a bent conformation when free in solution, with higher content of secondary structure and higher number of internal hydrogen bonds. TH-(1-43) in solution presents the highest mobility and least defined structure of all forms studied, and it shows an energetically more favorable interaction with membranes relative to THp-(1-43). Cationic residues, notably Arg15 and Arg16, which are the recognition sites of the kinases phosphorylating at Ser19, are also contributing to the interaction with the membrane. Our results reveal the structural flexibility of this region of TH, in accordance with the functional versatility and conformational adaptation to different partners. Furthermore, this structural information has potential relevance for the development of therapeutics for neurodegenerative disorders, through modulation of TH-partner interactions.
- Department of Biomedicine, University of Bergen, Jonas Lies vei 91, 5009 Bergen, Norway.
Organizational Affiliation: