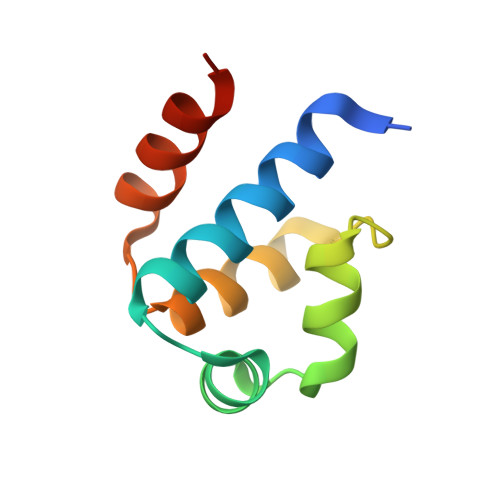
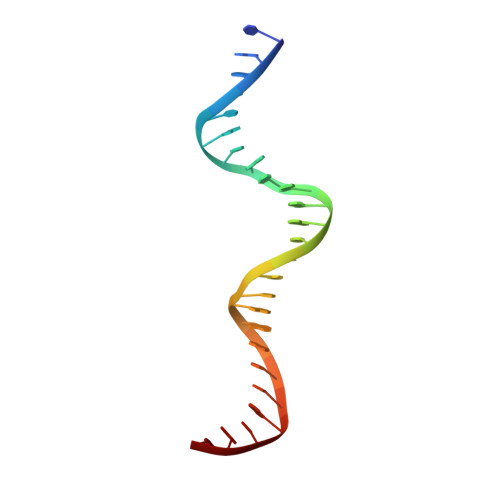
Structural analysis of DNA-protein complexes regulating the restriction-modification system Esp1396I.
Martin, R.N., McGeehan, J.E., Ball, N.J., Streeter, S.D., Thresh, S.J., Kneale, G.G.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 962-966
- PubMed: 23989141
- DOI: https://doi.org/10.1107/S174430911302126X
- Primary Citation of Related Structures:
4I8T, 4IWR - PubMed Abstract:
The controller protein of the type II restriction-modification (RM) system Esp1396I binds to three distinct DNA operator sequences upstream of the methyltransferase and endonuclease genes in order to regulate their expression. Previous biophysical and crystallographic studies have shown molecular details of how the controller protein binds to the operator sites with very different affinities. Here, two protein-DNA co-crystal structures containing portions of unbound DNA from native operator sites are reported. The DNA in both complexes shows significant distortion in the region between the conserved symmetric sequences, similar to that of a DNA duplex when bound by the controller protein (C-protein), indicating that the naked DNA has an intrinsic tendency to bend when not bound to the C-protein. Moreover, the width of the major groove of the DNA adjacent to a bound C-protein dimer is observed to be significantly increased, supporting the idea that this DNA distortion contributes to the substantial cooperativity found when a second C-protein dimer binds to the operator to form the tetrameric repression complex.
- Institute of Biomedical and Biomolecular Science, University of Portsmouth, King Henry I Street, Portsmouth, Hampshire PO1 2DY, England.
Organizational Affiliation: