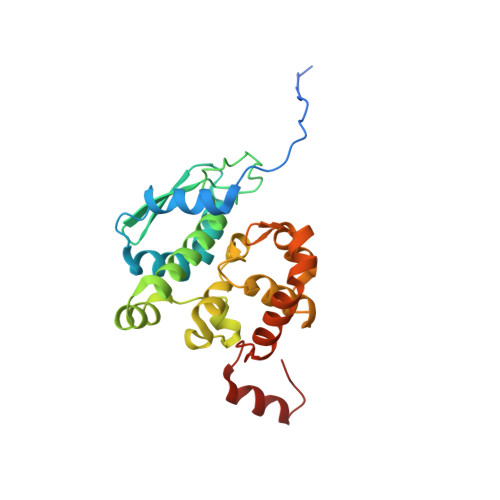
Bunyamwera virus possesses a distinct nucleocapsid protein to facilitate genome encapsidation
Li, B.B., Wang, Q., Pan, X., Fernandez de Castro, I., Sun, Y., Guo, Y., Tao, X., Risco, C., Sui, S.F., Lou, Z.Y.(2013) Proc Natl Acad Sci U S A 110: 9048-9053
- PubMed: 23569257
- DOI: https://doi.org/10.1073/pnas.1222552110
- Primary Citation of Related Structures:
4IJS - PubMed Abstract:
Bunyamwera virus (BUNV), which belongs to the genus Orthobunyavirus, is the prototypical virus of the Bunyaviridae family. Similar to other negative-sense single-stranded RNA viruses, bunyaviruses possess a nucleocapsid protein (NP) to facilitate genomic RNA encapsidation and virus replication. The structures of two NPs of members of different genera within the Bunyaviridae family have been reported. However, their structures, RNA-binding features, and functions beyond RNA binding significantly differ from one another. Here, we report the crystal structure of the BUNV NP-RNA complex. The polypeptide of the BUNV NP was found to possess a distinct fold among viral NPs. An N-terminal arm and a C-terminal tail were found to interact with neighboring NP protomers to form a tetrameric ring-shaped organization. Each protomer bound a 10-nt RNA molecule, which was acquired from the expression host, in the positively charged crevice between the N and C lobes. Inhomogeneous oligomerization was observed for the recombinant BUNV NP-RNA complex, which was similar to the Rift Valley fever virus NP-RNA complex. This result suggested that the flexibility of one NP protomer with adjacent protomers underlies the BUNV ribonucleoprotein complex (RNP) formation. Electron microscopy revealed that the monomer-sized NP-RNA complex was the building block of the natural BUNV RNP. Combined with previous results indicating that mutagenesis of the interprotomer or protein-RNA interface affects BUNV replication, our structure provides a great potential for understanding the mechanism underlying negative-sense single-stranded RNA RNP formation and enables the development of antiviral therapies targeting BUNV RNP formation.
- Laboratory of Structural Biology, School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: