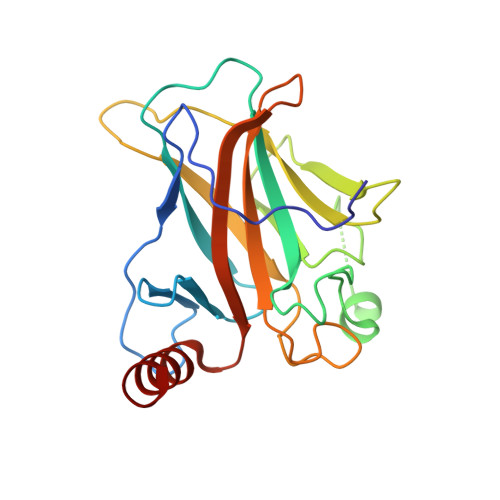
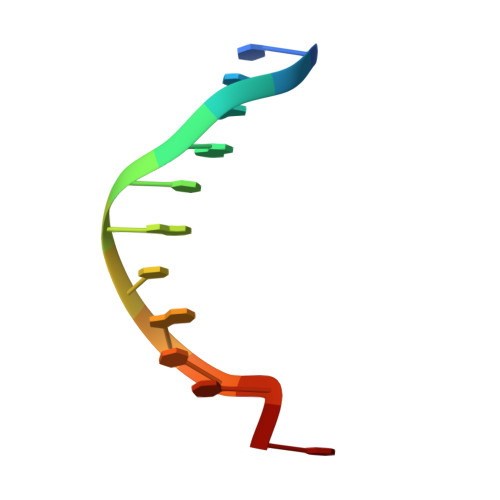
Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Eldar, A., Rozenberg, H., Diskin-Posner, Y., Rohs, R., Shakked, Z.(2013) Nucleic Acids Res 41: 8748-8759
- PubMed: 23863845
- DOI: https://doi.org/10.1093/nar/gkt630
- Primary Citation of Related Structures:
4IBQ, 4IBS, 4IBT, 4IBU, 4IBV, 4IBW, 4IBY, 4IBZ, 4IJT - PubMed Abstract:
A p53 hot-spot mutation found frequently in human cancer is the replacement of R273 by histidine or cysteine residues resulting in p53 loss of function as a tumor suppressor. These mutants can be reactivated by the incorporation of second-site suppressor mutations. Here, we present high-resolution crystal structures of the p53 core domains of the cancer-related proteins, the rescued proteins and their complexes with DNA. The structures show that inactivation of p53 results from the incapacity of the mutated residues to form stabilizing interactions with the DNA backbone, and that reactivation is achieved through alternative interactions formed by the suppressor mutations. Detailed structural and computational analysis demonstrates that the rescued p53 complexes are not fully restored in terms of DNA structure and its interface with p53. Contrary to our previously studied wild-type (wt) p53-DNA complexes showing non-canonical Hoogsteen A/T base pairs of the DNA helix that lead to local minor-groove narrowing and enhanced electrostatic interactions with p53, the current structures display Watson-Crick base pairs associated with direct or water-mediated hydrogen bonds with p53 at the minor groove. These findings highlight the pivotal role played by R273 residues in supporting the unique geometry of the DNA target and its sequence-specific complex with p53.
- Department of Structural Biology, Weizmann Institute of Science, Rehovot 76100, Israel and Molecular and Computational Biology Program, University of Southern California, Los Angeles, CA 90089, USA.
Organizational Affiliation: