Structure of human factor VIIa-soluble tissue factor with calcium, magnesium and rubidium
Vadivel, K., Schmidt, A., Cascio, D., Padmanabhan, K., Krishnaswamy, S., Brandstetter, H., Bajaj, S.P.(2021) Acta Crystallogr D Biol Crystallogr D77: 809-819
Experimental Data Snapshot
Starting Model: experimental
View more details
(2021) Acta Crystallogr D Biol Crystallogr D77: 809-819
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Coagulation factor VII | A [auth L] | 152 | Homo sapiens | Mutation(s): 0 EC: 3.4.21.21 | 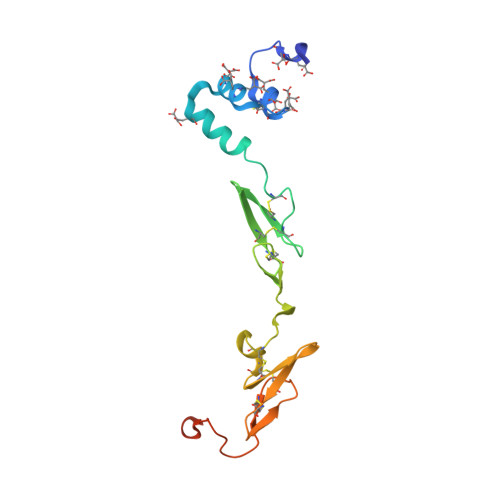 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P08709 (Homo sapiens) Explore P08709 Go to UniProtKB: P08709 | |||||
PHAROS: P08709 GTEx: ENSG00000057593 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P08709 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P08709-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Coagulation factor VII | B [auth H] | 254 | Homo sapiens | Mutation(s): 0 EC: 3.4.21.21 | 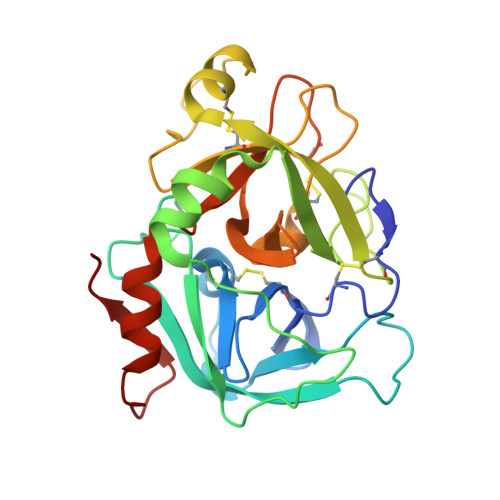 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P08709 (Homo sapiens) Explore P08709 Go to UniProtKB: P08709 | |||||
PHAROS: P08709 GTEx: ENSG00000057593 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P08709 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tissue factor | C [auth T] | 219 | Homo sapiens | Mutation(s): 0 | 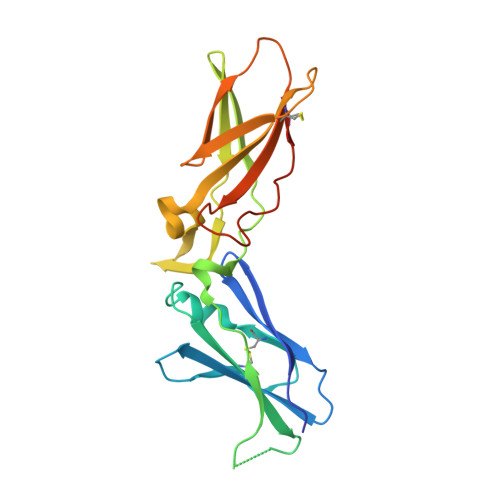 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P13726 (Homo sapiens) Explore P13726 Go to UniProtKB: P13726 | |||||
PHAROS: P13726 GTEx: ENSG00000117525 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13726 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BGC Query on BGC | D [auth L] | beta-D-glucopyranose C6 H12 O6 WQZGKKKJIJFFOK-VFUOTHLCSA-N |  | ||
| FUC Query on FUC | E [auth L] | alpha-L-fucopyranose C6 H12 O5 SHZGCJCMOBCMKK-SXUWKVJYSA-N |  | ||
| BEN Query on BEN | P [auth H] | BENZAMIDINE C7 H8 N2 PXXJHWLDUBFPOL-UHFFFAOYSA-N |  | ||
| RB Query on RB | H [auth L] J [auth L] N [auth L] O [auth L] S [auth H] | RUBIDIUM ION Rb NCCSSGKUIKYAJD-UHFFFAOYSA-N |  | ||
| CA Query on CA | G [auth L], K [auth L], M [auth L], Q [auth H] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | R [auth H], U [auth H], V [auth H], W [auth T] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | F [auth L], I [auth L], L | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CGU Query on CGU | A [auth L] | L-PEPTIDE LINKING | C6 H9 N O6 |  | GLU |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 69.9 | α = 90 |
| b = 81.06 | β = 90 |
| c = 126.42 | γ = 90 |
| Software Name | Purpose |
|---|---|
| ADSC | data collection |
| AMoRE | phasing |
| REFMAC | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |