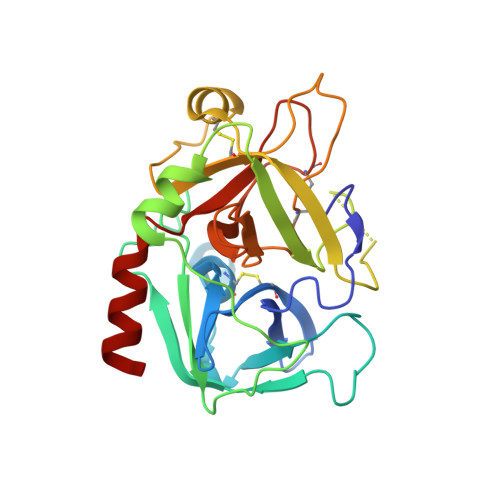
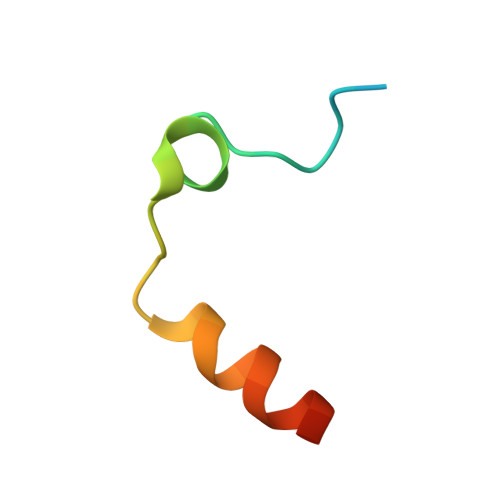
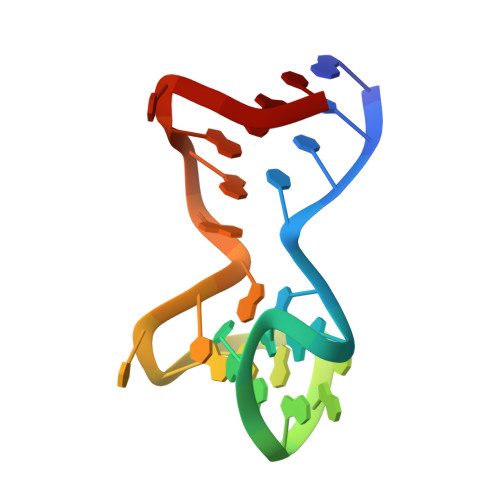
Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer.
Russo Krauss, I., Pica, A., Merlino, A., Mazzarella, L., Sica, F.(2013) Acta Crystallogr D Biol Crystallogr 69: 2403-2411
- PubMed: 24311581
- DOI: https://doi.org/10.1107/S0907444913022269
- Primary Citation of Related Structures:
4I7Y - PubMed Abstract:
Potent second-generation thrombin aptamers adopt a duplex-quadruplex bimodular folding and recognize thrombin exosite II with very high affinity and specificity. A sound model of these oligonucleotides, either free or in complex with thrombin, is not yet available. Here, a structural study of one of these aptamers, HD22-27mer, is presented. The crystal structure of this aptamer in complex with thrombin displays a novel architecture in which the helical stem is enchained to a pseudo-G-quadruplex. The results also underline the role of the residues that join the duplex and quadruplex motifs and control their recruitment in thrombin binding.
- Department of Chemical Sciences, University of Naples `Federico II', Complesso Universitario di Monte Sant'Angelo, I-80126 Naples, Italy.
Organizational Affiliation: