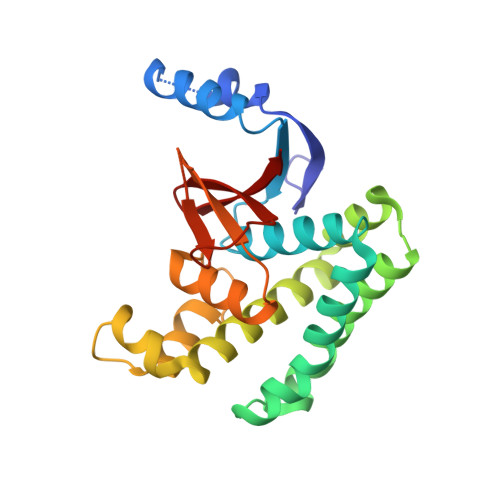
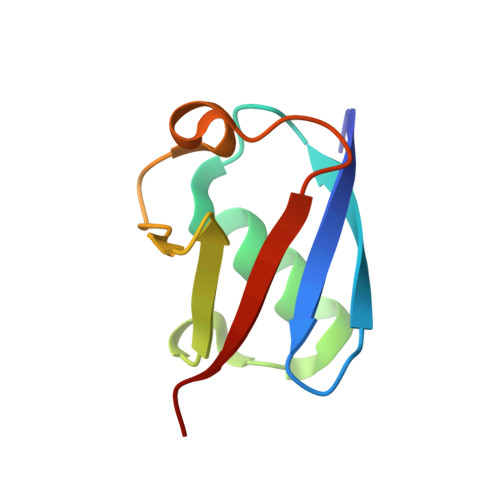
A strategy for modulation of enzymes in the ubiquitin system.
Ernst, A., Avvakumov, G., Tong, J., Fan, Y., Zhao, Y., Alberts, P., Persaud, A., Walker, J.R., Neculai, A.M., Neculai, D., Vorobyov, A., Garg, P., Beatty, L., Chan, P.K., Juang, Y.C., Landry, M.C., Yeh, C., Zeqiraj, E., Karamboulas, K., Allali-Hassani, A., Vedadi, M., Tyers, M., Moffat, J., Sicheri, F., Pelletier, L., Durocher, D., Raught, B., Rotin, D., Yang, J., Moran, M.F., Dhe-Paganon, S., Sidhu, S.S.(2013) Science 339: 590-595
- PubMed: 23287719
- DOI: https://doi.org/10.1126/science.1230161
- Primary Citation of Related Structures:
3I3T, 3MTN, 3N3K, 3V6E, 4I6L - PubMed Abstract:
The ubiquitin system regulates virtually all aspects of cellular function. We report a method to target the myriad enzymes that govern ubiquitination of protein substrates. We used massively diverse combinatorial libraries of ubiquitin variants to develop inhibitors of four deubiquitinases (DUBs) and analyzed the DUB-inhibitor complexes with crystallography. We extended the selection strategy to the ubiquitin conjugating (E2) and ubiquitin ligase (E3) enzymes and found that ubiquitin variants can also enhance enzyme activity. Last, we showed that ubiquitin variants can bind selectively to ubiquitin-binding domains. Ubiquitin variants exhibit selective function in cells and thus enable orthogonal modulation of specific enzymatic steps in the ubiquitin system.
- Terrence Donnelly Center for Cellular and Biomolecular Research, University of Toronto, 160 College Street, Toronto, Ontario, Canada.
Organizational Affiliation: