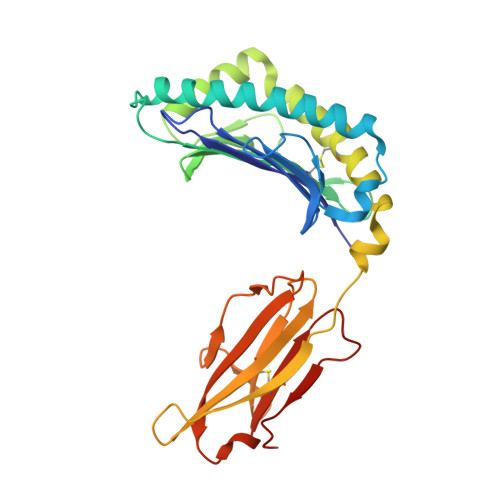
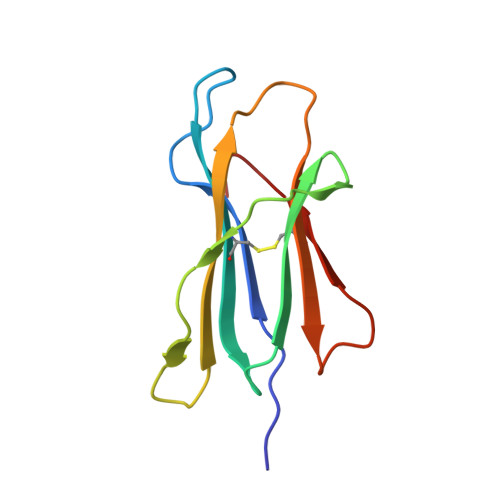
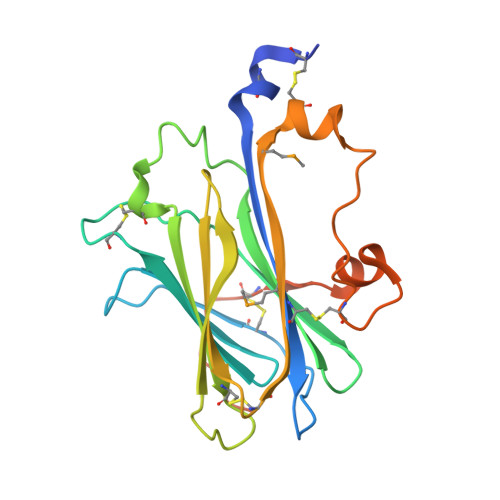
Structural Mechanism of ER Retrieval of MHC Class I by Cowpox.
McCoy, W.H., Wang, X., Yokoyama, W.M., Hansen, T.H., Fremont, D.H.(2012) PLoS Biol 10: e1001432-e1001432
- PubMed: 23209377
- DOI: https://doi.org/10.1371/journal.pbio.1001432
- Primary Citation of Related Structures:
4HKJ - PubMed Abstract:
One of the hallmarks of viral immune evasion is the capacity to disrupt major histocompatibility complex class I (MHCI) antigen presentation to evade T-cell detection. Cowpox virus encoded protein CPXV203 blocks MHCI surface expression by exploiting the KDEL-receptor recycling pathway, and here we show that CPXV203 directly binds a wide array of fully assembled MHCI proteins, both classical and non-classical. Further, the stability of CPXV203/MHCI complexes is highly pH dependent, with dramatically increased affinities at the lower pH of the Golgi relative to the endoplasmic reticulum (ER). Crystallographic studies reveal that CPXV203 adopts a beta-sandwich fold similar to poxvirus chemokine binding proteins, and binds the same highly conserved MHCI determinants located under the peptide-binding platform that tapasin, CD8, and natural killer (NK)-receptors engage. Mutagenesis of the CPXV203/MHCI interface identified the importance of two CPXV203 His residues that confer low pH stabilization of the complex and are critical to ER retrieval of MHCI. These studies clarify mechanistically how CPXV203 coordinates with other cowpox proteins to thwart antigen presentation.
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, Missouri, United States of America.
Organizational Affiliation: