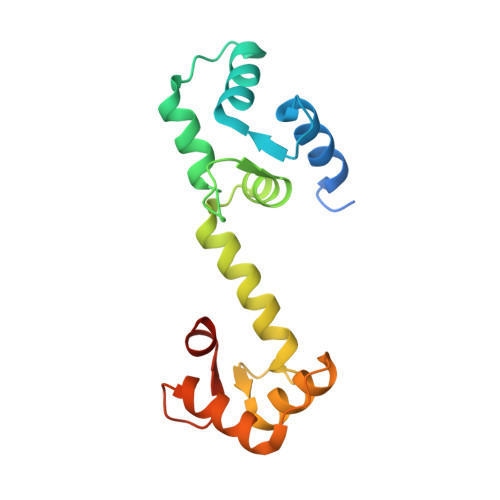
A novel trans conformation of ligand-free calmodulin
Kumar, V., Chichili, V.P.R., Tang, X., Sivaraman, J.(2013) PLoS One 8: e54834-e54834
- PubMed: 23382982
- DOI: https://doi.org/10.1371/journal.pone.0054834
- Primary Citation of Related Structures:
4HEX - PubMed Abstract:
Calmodulin (CaM) is a highly conserved eukaryotic protein that binds specifically to more than 100 target proteins in response to calcium (Ca(2+)) signal. CaM adopts a considerable degree of structural plasticity to accomplish this physiological role; however, the nature and extent of this plasticity remain to be fully understood. Here, we report the crystal structure of a novel trans conformation of ligand-free CaM where the relative disposition of two lobes of CaM is different, a conformation to-date not reported. While no major structural changes were observed in the independent N- and C-lobes as compared with previously reported structures of Ca(2+)/CaM, the central helix was tilted by ~90° at Arg75. This is the first crystal structure of CaM to show a drastic conformational change in the central helix, and reveals one of several possible conformations of CaM to engage with its binding partner.
- Department of Biological Sciences, National University of Singapore, Republic of Singapore, Republic of Singapore.
Organizational Affiliation: