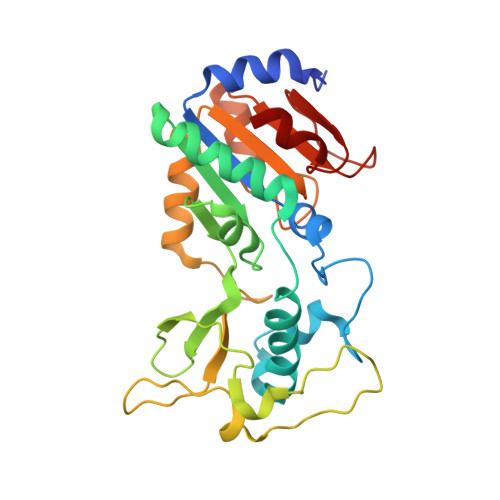
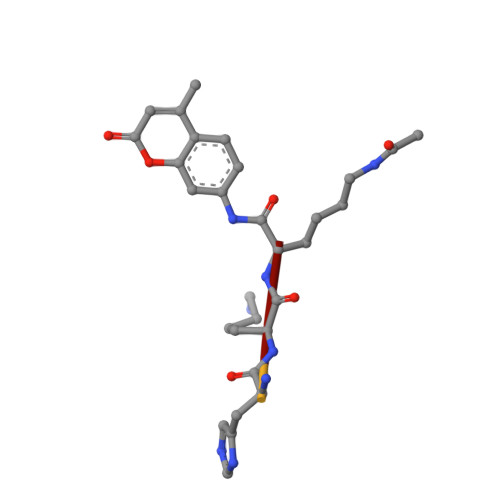
A molecular mechanism for direct sirtuin activation by resveratrol.
Gertz, M., Nguyen, G.T., Fischer, F., Suenkel, B., Schlicker, C., Franzel, B., Tomaschewski, J., Aladini, F., Becker, C., Wolters, D., Steegborn, C.(2012) PLoS One 7: e49761-e49761
- PubMed: 23185430
- DOI: https://doi.org/10.1371/journal.pone.0049761
- Primary Citation of Related Structures:
4HD8, 4HDA - PubMed Abstract:
Sirtuins are protein deacetylases regulating metabolism, stress responses, and aging processes, and they were suggested to mediate the lifespan extending effect of a low calorie diet. Sirtuin activation by the polyphenol resveratrol can mimic such lifespan extending effects and alleviate metabolic diseases. The mechanism of Sirtuin stimulation is unknown, hindering the development of improved activators. Here we show that resveratrol inhibits human Sirt3 and stimulates Sirt5, in addition to Sirt1, against fluorophore-labeled peptide substrates but also against peptides and proteins lacking the non-physiological fluorophore modification. We further present crystal structures of Sirt3 and Sirt5 in complex with fluorogenic substrate peptide and modulator. The compound acts as a top cover, closing the Sirtuin's polypeptide binding pocket and influencing details of peptide binding by directly interacting with this substrate. Our results provide a mechanism for the direct activation of Sirtuins by small molecules and suggest that activators have to be tailored to a specific Sirtuin/substrate pair.
- Department of Biochemistry, University of Bayreuth, Bayreuth, Germany.
Organizational Affiliation: