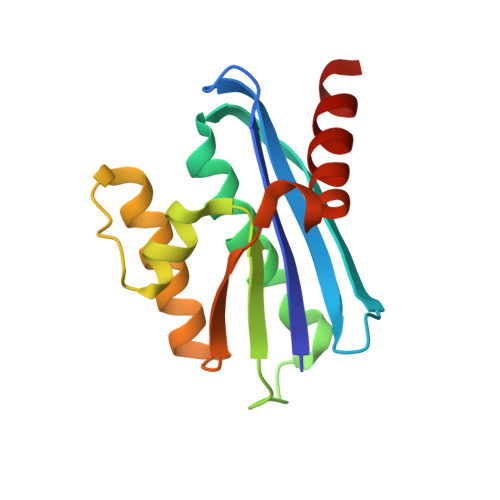
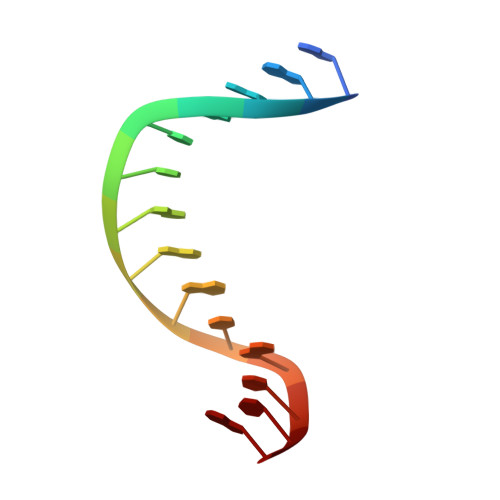
Crystal structure of metagenome-derived LC11-RNase H1 in complex with RNA/DNA hybrid
Nguyen, T.N., You, D.J., Matsumoto, H., Kanaya, E., Koga, Y., Kanaya, S.(2013) J Struct Biol 182: 144-154
- PubMed: 23500886
- DOI: https://doi.org/10.1016/j.jsb.2013.02.018
- Primary Citation of Related Structures:
4H8K - PubMed Abstract:
LC11-RNase H1 is a Sulfolobus tokodaii RNase H1 (Sto-RNase H1) homologue isolated by metagenomic approach. In this study, the crystal structure of LC11-RNase H1 in complex with an RNA/DNA substrate was determined. Unlike Bacillus halodurans RNase H1 without hybrid binding domain (HBD) (Bh-RNase HC) and human RNase H1 without HBD (Hs-RNase HC), LC11-RNase H1 interacts with four non-consecutive 2'-OH groups of the RNA strand. The lack of interactions with four consecutive 2'-OH groups leads to a dramatic decrease in the ability of LC11-RNase H1 to cleave the DNA-RNA-DNA/DNA substrate containing four ribonucleotides as compared to those to cleave the substrates containing five and six ribonucleotides. The interaction of LC11-RNase H1 with the DNA strand is also different from those of Bh-RNase HC and Hs-RNase HC. Beside the common phosphate-binding pocket, LC11-RNase H1 has a unique DNA-binding channel. Furthermore, the active-site residues of LC11-RNase H1 are located farther away from the scissile phosphate group than those of Bh-RNase HC and Hs-RNase HC. Modeling of Sto-RNase H1 in complex with the 14bp RNA/DNA substrate, together with the structure-based mutational analyses, suggest that the ability of Sto-RNase H1 to cleave double-stranded RNA is dependent on the local conformation of the basic residues located at the DNA binding site.
- Department of Material and Life Science, Graduate School of Engineering, Osaka University, 2-1 Yamadaoka, Suita, Osaka 565-0871, Japan.
Organizational Affiliation: