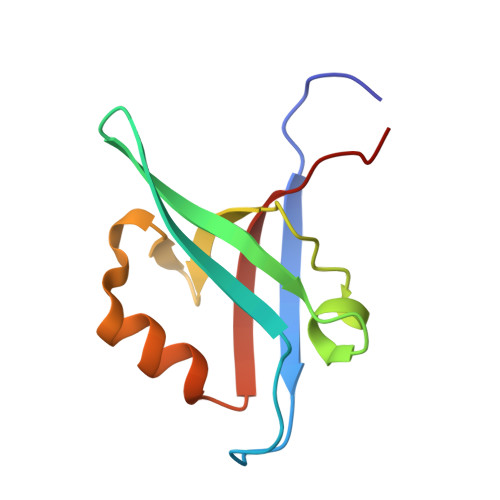
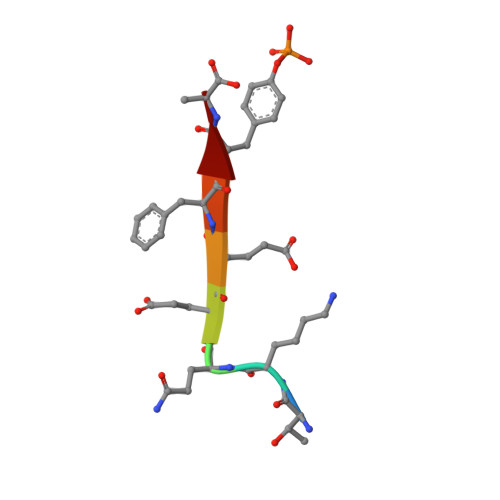
The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics.
Liu, X., Shepherd, T.R., Murray, A.M., Xu, Z., Fuentes, E.J.(2013) Structure 21: 342-354
- PubMed: 23395182
- DOI: https://doi.org/10.1016/j.str.2013.01.004
- Primary Citation of Related Structures:
4GVC, 4GVD - PubMed Abstract:
PDZ (PSD-95/Dlg/ZO-1) domains are protein-protein interaction modules often regulated by ligand phosphorylation. Here, we investigated the specificity, structure, and dynamics of Tiam1 PDZ domain/ligand interactions. We show that the PDZ domain specifically binds syndecan1 (SDC1), phosphorylated SDC1 (pSDC1), and SDC3 but not other syndecan isoforms. The crystal structure of the PDZ/SDC1 complex indicates that syndecan affinity is derived from amino acids beyond the four C-terminal residues. Remarkably, the crystal structure of the PDZ/pSDC1 complex reveals a binding pocket that accommodates the phosphoryl group. Methyl relaxation experiments of PDZ/SCD1 and PDZ/pSDC1 complexes reveal that PDZ-phosphoryl interactions dampen dynamic motions in a distal region of the PDZ domain by decoupling them from the ligand-binding site. Our data are consistent with a selection model by which specificity and phosphorylation regulate PDZ/syndecan interactions and signaling events. Importantly, our relaxation data demonstrate that PDZ/phospho-ligand interactions regulate protein dynamics and their coupling to distal sites.
- Department of Biochemistry, University of Iowa, Carver College of Medicine, Iowa City, IA 52242, USA.
Organizational Affiliation: