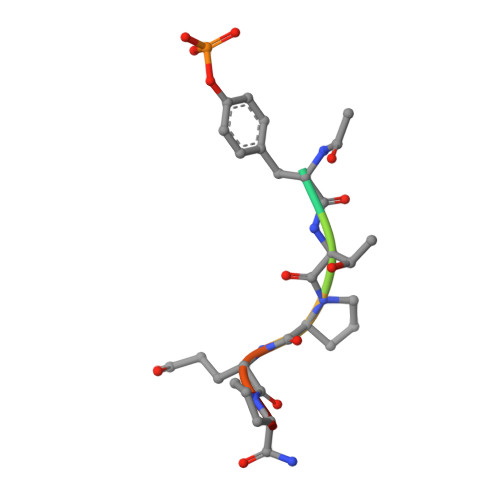
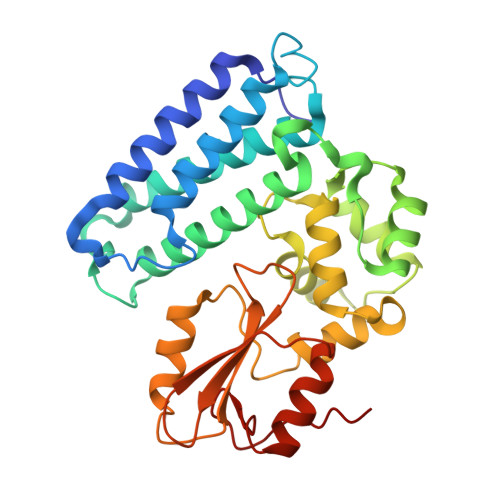
The paradox of conformational constraint in the design of Cbl(TKB)-binding peptides.
Kumar, E.A., Chen, Q., Kizhake, S., Kolar, C., Kang, M., Chang, C.E., Borgstahl, G.E., Natarajan, A.(2013) Sci Rep 3: 1639-1639
- PubMed: 23572190
- DOI: https://doi.org/10.1038/srep01639
- Primary Citation of Related Structures:
4GPL - PubMed Abstract:
Solving the crystal structure of Cbl(TKB) in complex with a pentapeptide, pYTPEP, revealed that the PEP region adopted a poly-L-proline type II (PPII) helix. An unnatural amino acid termed a proline-templated glutamic acid (ptE) that constrained both the backbone and sidechain to the bound conformation was synthesized and incorporated into the pYTPXP peptide. We estimated imposing structural constraints onto the backbone and sidechain of the peptide and preorganize it to the bound conformation in solution will yield nearly an order of magnitude improvement in activity. NMR studies confirmed that the ptE-containing peptide adopts the PPII conformation, however, competitive binding studies showed an order of magnitude loss of activity. Given the emphasis that is placed on imposing structural constraints, we provide an example to support the contrary. These results point to conformational flexibility at the interface, which have implications in the design of potent Cbl(TKB)-binding peptides.
- Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, Nebraska 68022, United States.
Organizational Affiliation: