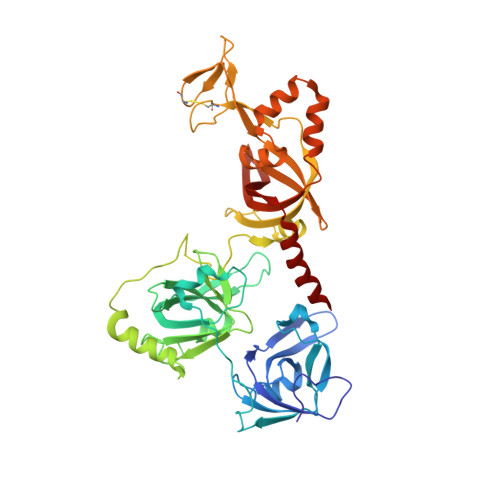
Structure and conformational change of a replication protein A heterotrimer bound to ssDNA.
Fan, J., Pavletich, N.P.(2012) Genes Dev 26: 2337-2347
- PubMed: 23070815
- DOI: https://doi.org/10.1101/gad.194787.112
- Primary Citation of Related Structures:
4GOP - PubMed Abstract:
Replication protein A (RPA) is the main eukaryotic ssDNA-binding protein with essential roles in DNA replication, recombination, and repair. RPA maintains the DNA as single-stranded and also interacts with other DNA-processing proteins, coordinating their assembly and disassembly on DNA. RPA binds to ssDNA in two conformational states with opposing affinities for DNA and proteins. The RPA-protein interactions are compatible with a low DNA affinity state that involves DNA-binding domain A (DBD-A) and DBD-B but not with the high DNA affinity state that additionally engages DBD-C and DBD-D. The structure of the high-affinity RPA-ssDNA complex reported here shows a compact quaternary structure held together by a four-way interface between DBD-B, DBD-C, the intervening linker (BC linker), and ssDNA. The BC linker binds into the DNA-binding groove of DBD-B, mimicking DNA. The associated conformational change and partial occlusion of the DBD-A-DBA-B protein-protein interaction site establish a mechanism for the allosteric coupling of RPA-DNA and RPA-protein interactions.
- Sloan-Kettering Division, Joan and Sanford I. Weill Graduate School of Medical Sciences, Cornell University, New York, New York 10065, USA.
Organizational Affiliation: