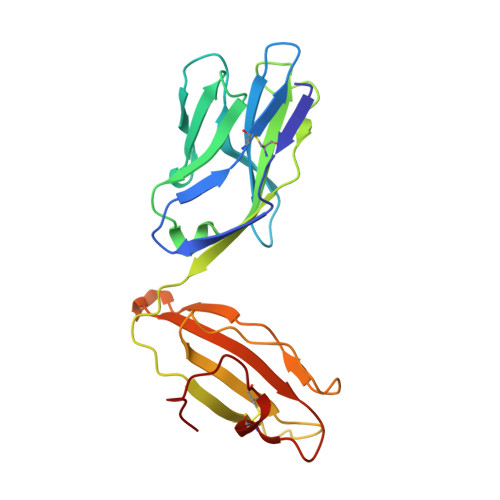
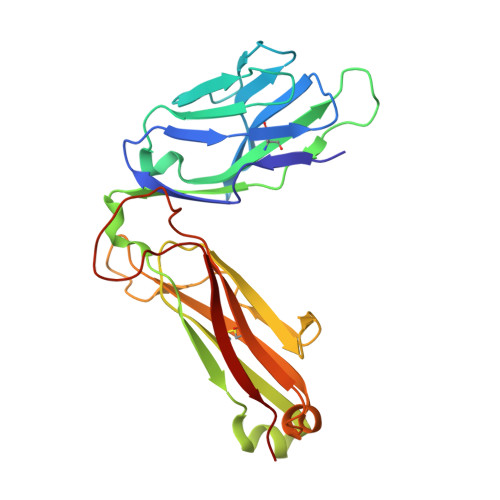
Minimal conformational plasticity enables TCR cross-reactivity to different MHC class II heterodimers.
Holland, C.J., Rizkallah, P.J., Vollers, S., Calvo-Calle, J.M., Madura, F., Fuller, A., Sewell, A.K., Stern, L.J., Godkin, A., Cole, D.K.(2012) Sci Rep 2: 629-629
- PubMed: 22953050
- DOI: https://doi.org/10.1038/srep00629
- Primary Citation of Related Structures:
4GKZ - PubMed Abstract:
Successful immunity requires that a limited pool of αβ T-cell receptors (TCRs) provide cover for a vast number of potential foreign peptide antigens presented by 'self' major histocompatibility complex (pMHC) molecules. Structures of unligated and ligated MHC class-I-restricted TCRs with different ligands, supplemented with biophysical analyses, have revealed a number of important mechanisms that govern TCR mediated antigen recognition. HA1.7 TCR binding to the influenza hemagglutinin antigen (HA(306-318)) presented by HLA-DR1 or HLA-DR4 represents an ideal system for interrogating pMHC-II antigen recognition. Accordingly, we solved the structure of the unligated HA1.7 TCR and compared it to both complex structures. Despite a relatively rigid binding mode, HA1.7 T-cells could tolerate mutations in key contact residues within the peptide epitope. Thermodynamic analysis revealed that limited plasticity and extreme favorable entropy underpinned the ability of the HA1.7 T-cell clone to cross-react with HA(306-318) presented by multiple MHC-II alleles.
- Institute of Infection and Immunity, Cardiff University School of Medicine, Cardiff, United Kingdom.
Organizational Affiliation: