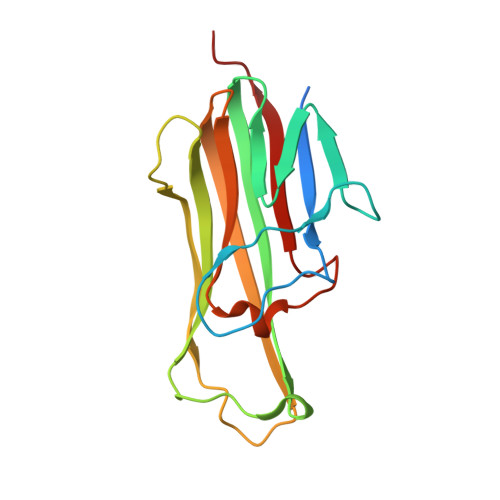
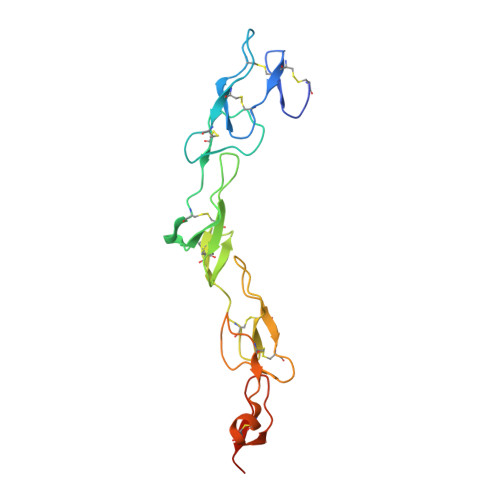
RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor.
Nelson, C.A., Warren, J.T., Wang, M.W., Teitelbaum, S.L., Fremont, D.H.(2012) Structure 20: 1971-1982
- PubMed: 23039992
- DOI: https://doi.org/10.1016/j.str.2012.08.030
- Primary Citation of Related Structures:
4E4D, 4GIQ - PubMed Abstract:
Osteoprotegerin (OPG) and receptor activator of nuclear factor κB (RANK) are members of the tumor necrosis factor receptor (TNFR) superfamily that regulate osteoclast formation and function by competing for RANK ligand (RANKL). RANKL promotes osteoclast development through RANK activation, while OPG inhibits this process by sequestering RANKL. For comparison, we solved crystal structures of RANKL with RANK and RANKL with OPG. Complementary biochemical and functional studies reveal that the monomeric cytokine-binding region of OPG binds RANKL with ∼500-fold higher affinity than RANK and inhibits RANKL-stimulated osteoclastogenesis ∼150 times more effectively, in part because the binding cleft of RANKL makes unique contacts with OPG. Several side chains as well as the C-D and D-E loops of RANKL occupy different orientations when bound to OPG versus RANK. High affinity OPG binding requires a 90s loop Phe residue that is mutated in juvenile Paget's disease. These results suggest cytokine plasticity may help to fine-tune specific tumor necrosis factor (TNF)-family cytokine/receptor pair selectivity.
- Department of Pathology and Immunology, Washington University School of Medicine, 660 S. Euclid Avenue, St. Louis, MO 63110-1093, USA.
Organizational Affiliation: