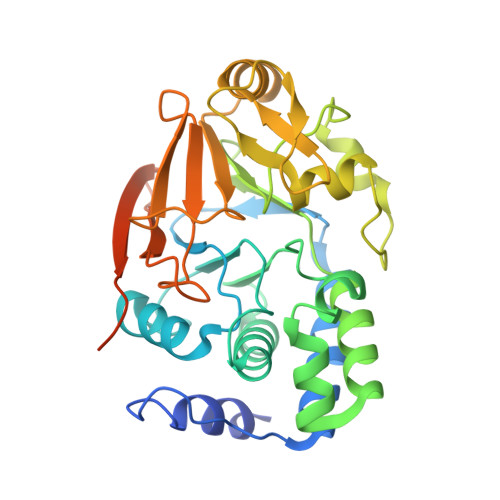
Development of a Peptide that Selectively Activates Protein Phosphatase-1 in Living Cells.
Chatterjee, J., Beullens, M., Sukackaite, R., Qian, J., Lesage, B., Hart, D.J., Bollen, M., Kohn, M.(2012) Angew Chem Int Ed Engl 51: 10054-10059
- PubMed: 22962028
- DOI: https://doi.org/10.1002/anie.201204308
- Primary Citation of Related Structures:
4G9J - PubMed Abstract:
that activates protein phosphatase-1 (PP1) by disrupting a subset of PP1 complexes in living cells has been developed. Activated PP1 rapidly dephosphorylates its substrates, counteracting kinase activity inside cells. Activation of PP1 can thus be a novel approach to study PP1 function and to counteract Ser/Thr kinase activity under pathologically increased kinase signaling.
- Genome Biology Unit, EMBL, Meyerhofstrasse 1, 69117 Heidelberg, Germany.
Organizational Affiliation: