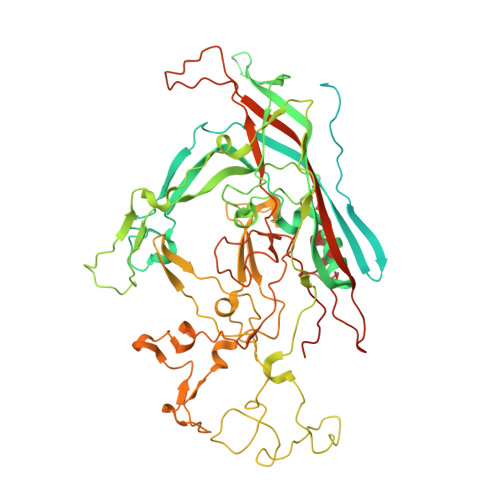
Structural characterization of h-1 parvovirus: comparison of infectious virions to empty capsids.
Halder, S., Nam, H.J., Govindasamy, L., Vogel, M., Dinsart, C., Salome, N., McKenna, R., Agbandje-McKenna, M.(2013) J Virol 87: 5128-5140
- PubMed: 23449783
- DOI: https://doi.org/10.1128/JVI.03416-12
- Primary Citation of Related Structures:
4G0R, 4GBT - PubMed Abstract:
The structure of single-stranded DNA (ssDNA) packaging H-1 parvovirus (H-1PV), which is being developed as an antitumor gene delivery vector, has been determined for wild-type (wt) virions and noninfectious (empty) capsids to 2.7- and 3.2-Å resolution, respectively, using X-ray crystallography. The capsid viral protein (VP) structure consists of an α-helix and an eight-stranded anti-parallel β-barrel with large loop regions between the strands. The β-barrel and loops form the capsid core and surface, respectively. In the wt structure, 600 nucleotides are ordered in an interior DNA binding pocket of the capsid. This accounts for ∼12% of the H-1PV genome. The wt structure is identical to the empty capsid structure, except for side chain conformation variations at the nucleotide binding pocket. Comparison of the H-1PV nucleotides to those observed in canine parvovirus and minute virus of mice, two members of the genus Parvovirus, showed both similarity in structure and analogous interactions. This observation suggests a functional role, such as in capsid stability and/or ssDNA genome recognition for encapsulation. The VP structure differs from those of other parvoviruses in surface loop regions that control receptor binding, tissue tropism, pathogenicity, and antibody recognition, including VP sequences reported to determine tumor cell tropism for oncotropic rodent parvoviruses. These structures of H-1PV provide insight into structural features that dictate capsid stabilization following genome packaging and three-dimensional information applicable for rational design of tumor-targeted recombinant gene delivery vectors.
- Department of Biochemistry and Molecular Biology, University of Florida, Gainesville, Florida, USA.
Organizational Affiliation: