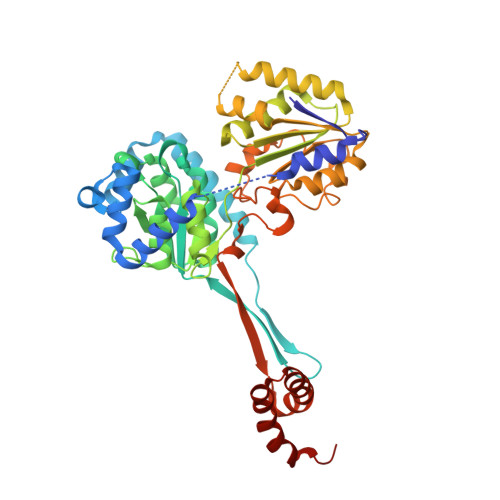
Structural basis for the rational design of new anti-Brucella agents: The crystal structure of the C366S mutant of l-histidinol dehydrogenase from Brucella suis.
D'ambrosio, K., Lopez, M., Dathan, N.A., Ouahrani-Bettache, S., Kohler, S., Ascione, G., Monti, S.M., Winum, J.Y., De Simone, G.(2014) Biochimie 97: 114-120
- PubMed: 24140957
- DOI: https://doi.org/10.1016/j.biochi.2013.09.028
- Primary Citation of Related Structures:
4G07, 4G09 - PubMed Abstract:
L-Histidinol dehydrogenase from Brucella suis (BsHDH) is an enzyme involved in the histidine biosynthesis pathway which is absent in mammals, thus representing a very interesting target for the development of anti-Brucella agents. In this paper we report the crystallographic structure of a mutated form of BsHDH both in its unbound form and in complex with a nanomolar inhibitor. These studies provide the first structural background for the rational design of potent HDH inhibitors, thus offering new hints for clinical applications.
- Istituto di Biostrutture e Bioimmagini-CNR, Via Mezzocannone 16, 80134 Napoli, Italy.
Organizational Affiliation: