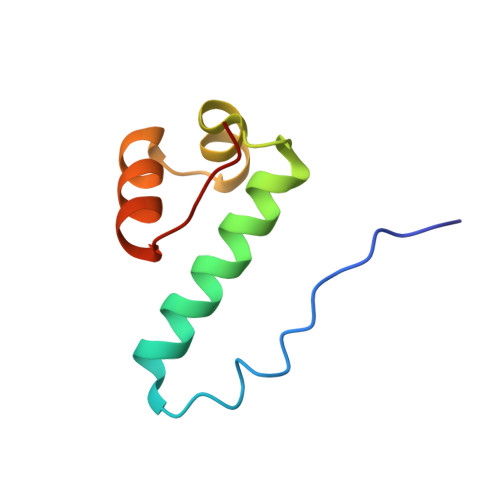
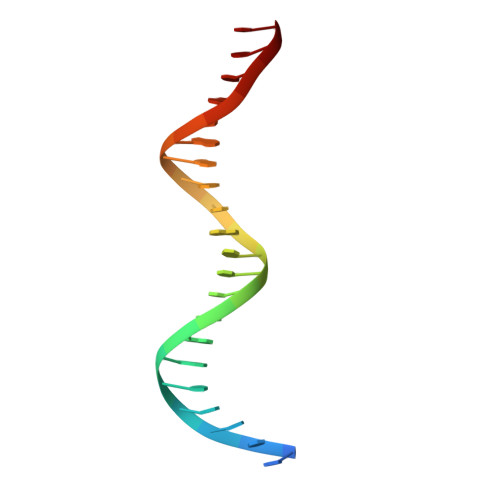
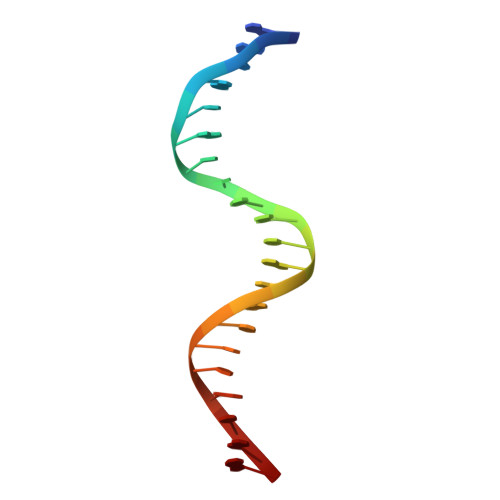
DNA Recognition by a sigma (54) Transcriptional Activator from Aquifex aeolicus.
Vidangos, N.K., Heideker, J., Lyubimov, A., Lamers, M., Huo, Y., Pelton, J.G., Ton, J., Gralla, J., Berger, J., Wemmer, D.E.(2014) J Mol Biology 426: 3553-3568
- PubMed: 25158097
- DOI: https://doi.org/10.1016/j.jmb.2014.08.009
- Primary Citation of Related Structures:
4FTH - PubMed Abstract:
Transcription initiation by bacterial σ(54)-polymerase requires the action of a transcriptional activator protein. Activators bind sequence-specifically upstream of the transcription initiation site via a DNA-binding domain (DBD). The structurally characterized DBDs from activators all belong to the Fis (factor for inversion stimulation) family of helix-turn-helix DNA-binding proteins. We report here structures of the free and DNA-bound forms of the DBD of NtrC4 (4DBD) from Aquifex aeolicus, a member of the NtrC family of σ(54) activators. Two NtrC4-binding sites were identified upstream (-145 and -85bp) from the start of the lpxC gene, which is responsible for the first committed step in lipid A biosynthesis. This is the first experimental evidence for σ(54) regulation in lpxC expression. 4DBD was crystallized both without DNA and in complex with the -145-binding site. The structures, together with biochemical data, indicate that NtrC4 binds to DNA in a manner that is similar to that of its close homolog, Fis. The greater sequence specificity for the binding of 4DBD relative to Fis seems to arise from a larger number of base-specific contacts contributing to affinity than for Fis.
- Department of Chemistry, University of California, MC-1460, Berkeley, CA 94720, USA.
Organizational Affiliation: