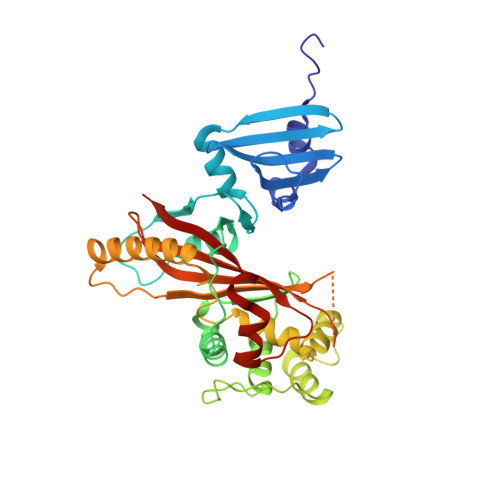
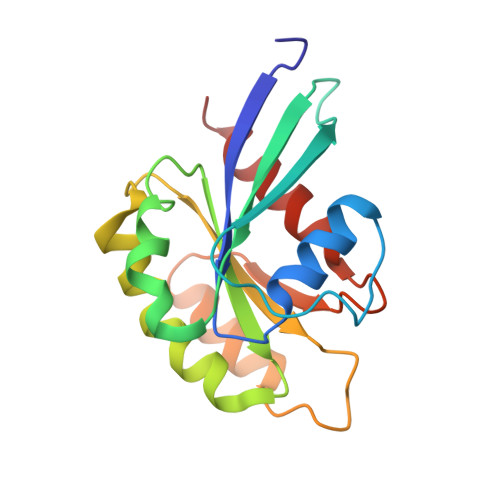
Structurally Distinct Bacterial TBC-like GAPs Link Arf GTPase to Rab1 Inactivation to Counteract Host Defenses.
Dong, N., Zhu, Y., Lu, Q., Hu, L., Zheng, Y., Shao, F.(2012) Cell 150: 1029-1041
- PubMed: 22939626
- DOI: https://doi.org/10.1016/j.cell.2012.06.050
- Primary Citation of Related Structures:
4FMA, 4FMB, 4FMC, 4FMD, 4FME - PubMed Abstract:
Rab GTPases are frequent targets of vacuole-living bacterial pathogens for appropriate trafficking of the vacuole. Here we discover that bacterial effectors including VirA from nonvacuole Shigella flexneri and EspG from extracellular Enteropathogenic Escherichia coli (EPEC) harbor TBC-like dual-finger motifs and exhibits potent RabGAP activities. Specific inactivation of Rab1 by VirA/EspG disrupts ER-to-Golgi trafficking. S. flexneri intracellular persistence requires VirA TBC-like GAP activity that mediates bacterial escape from autophagy-mediated host defense. Rab1 inactivation by EspG severely blocks host secretory pathway, resulting in inhibited interleukin-8 secretion from infected cells. Crystal structures of VirA/EspG-Rab1-GDP-aluminum fluoride complexes highlight TBC-like catalytic role for the arginine and glutamine finger residues and reveal a 3D architecture distinct from that of the TBC domain. Structure of Arf6-EspG-Rab1 ternary complex illustrates a pathogenic signaling complex that rewires host Arf signaling to Rab1 inactivation. Structural distinctions of VirA/EspG further predict a possible extensive presence of TBC-like RabGAP effectors in counteracting various host defenses.
- National Institute of Biological Sciences, Beijing 102206, China.
Organizational Affiliation: