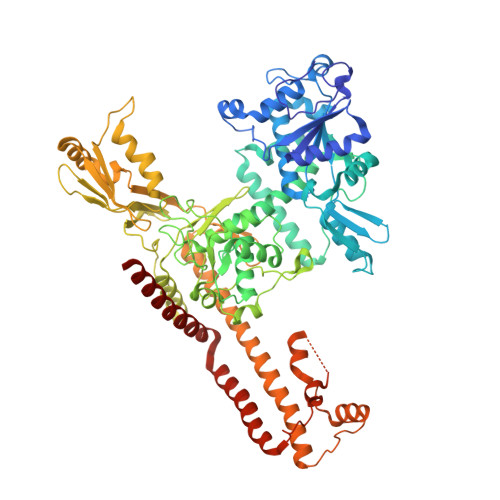
The Structure of DNA-Bound Human Topoisomerase II Alpha: Conformational Mechanisms for Coordinating Inter-Subunit Interactions with DNA Cleavage.
Wendorff, T.J., Schmidt, B.H., Heslop, P., Austin, C.A., Berger, J.M.(2012) J Mol Biology 424: 109-124
- PubMed: 22841979
- DOI: https://doi.org/10.1016/j.jmb.2012.07.014
- Primary Citation of Related Structures:
4FM9 - PubMed Abstract:
Type II topoisomerases are required for the management of DNA superhelicity and chromosome segregation, and serve as frontline targets for a variety of small-molecule therapeutics. To better understand how these enzymes act in both contexts, we determined the 2.9-Å-resolution structure of the DNA cleavage core of human topoisomerase IIα (TOP2A) bound to a doubly nicked, 30-bp duplex oligonucleotide. In accord with prior biochemical and structural studies, TOP2A significantly bends its DNA substrate using a bipartite, nucleolytic center formed at an N-terminal dimerization interface of the cleavage core. However, the protein also adopts a global conformation in which the second of its two inter-protomer contact points, one at the C-terminus, has separated. This finding, together with comparative structural analyses, reveals that the principal site of DNA engagement undergoes highly quantized conformational transitions between distinct binding, cleavage, and drug-inhibited states that correlate with the control of subunit-subunit interactions. Additional consideration of our TOP2A model in light of an etoposide-inhibited complex of human topoisomerase IIβ (TOP2B) suggests possible modification points for developing paralog-specific inhibitors to overcome the tendency of topoisomerase II-targeting chemotherapeutics to generate secondary malignancies.
- Biophysics Graduate Program, University of California Berkeley, Berkeley, CA 94720, USA.
Organizational Affiliation: