Molecular Recognition of Canonical and Deaminated Bases by P. abyssi Family B DNA Polymerase.
Gouge, J., Ralec, C., Henneke, G., Delarue, M.(2012) J Mol Biology 423: 315-336
- PubMed: 22902479
- DOI: https://doi.org/10.1016/j.jmb.2012.07.025
- Primary Citation of Related Structures:
4FLT, 4FLU, 4FLV, 4FLW, 4FLX, 4FLY, 4FLZ, 4FM0, 4FM1, 4FM2 - PubMed Abstract:
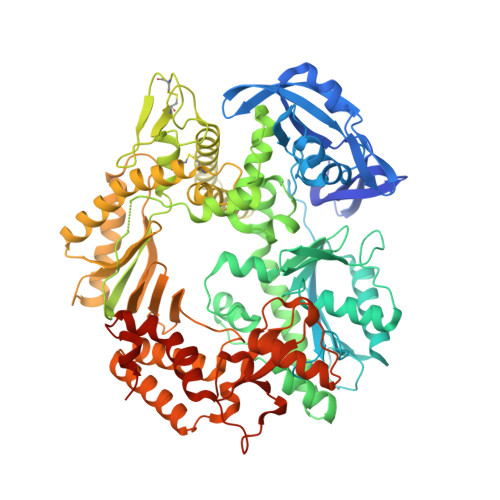
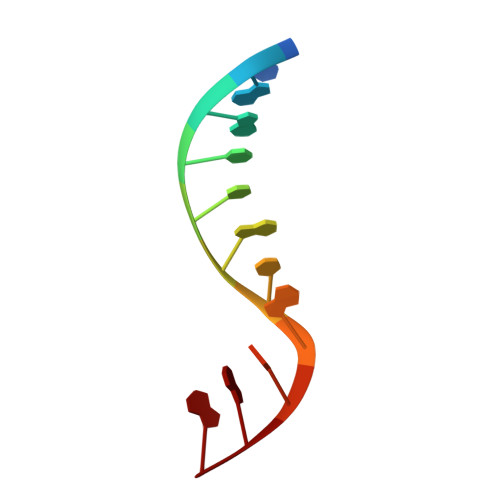
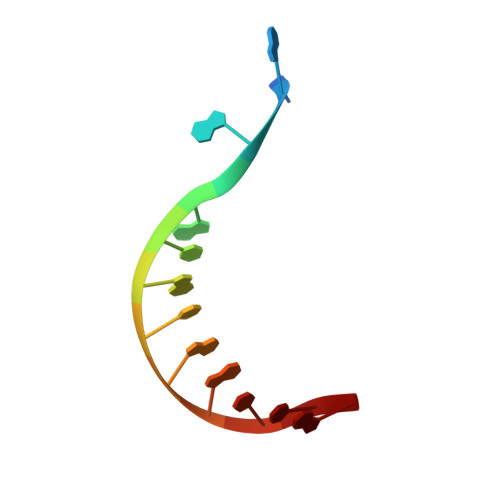
Euryarchaeal polymerase B can recognize deaminated bases on the template strand, effectively stalling the replication fork 4nt downstream the modified base. Using Pyrococcus abyssi DNA B family polymerase (PabPolB), we investigated the discrimination between deaminated and natural nucleotide(s) by primer extension assays, electrophoretic mobility shift assays, and X-ray crystallography. Structures of complexes between the protein and DNA duplexes with either a dU or a dH in position +4 were solved at 2.3Å and 2.9Å resolution, respectively. The PabPolB is found in the editing mode. A new metal binding site has been uncovered below the base-checking cavity where the +4 base is flipped out; it is fully hydrated in an octahedral fashion and helps guide the strongly kinked template strand. Four other crystal structures with each of the canonical bases were also solved in the editing mode, and the presence of three nucleotides in the exonuclease site caused a shift in the coordination state of its metal A from octahedral to tetrahedral. Surprisingly, we find that all canonical bases also enter the base-checking pocket with very small differences in the binding geometry and in the calculated binding free energy compared to deaminated ones. To explain how this can lead to stalling of the replication fork, the full catalytic pathway and its branches must be taken into account, during which the base is checked several times. Our results strongly suggest a switch from elongation to editing modes right after nucleotide insertion when the modified base is at position +5.
- Unité de Dynamique Structurale des Macromolécules, UMR 3528 du CNRS, Institut Pasteur, 25 rue du Dr Roux, 75015 Paris, France.
Organizational Affiliation: