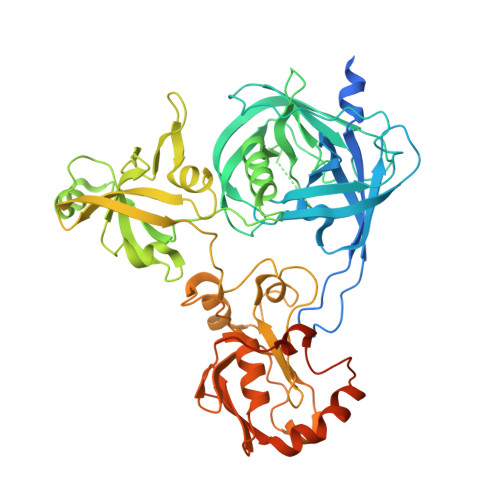
Crystal structure of Arabidopsis deg2 protein reveals an internal PDZ ligand locking the hexameric resting state.
Sun, R., Fan, H., Gao, F., Lin, Y., Zhang, L., Gong, W., Liu, L.(2012) J Biological Chem 287: 37564-37569
- PubMed: 22961982
- DOI: https://doi.org/10.1074/jbc.M112.394585
- Primary Citation of Related Structures:
4FLN - PubMed Abstract:
Eukaryotic organelles have developed elaborate protein quality control systems to ensure their normal activity, among which Deg/HtrA proteases play an essential role. Plant Deg2 protease is a homologue of prokaryotic DegQ/DegP proteases and is located in the chloroplast stroma, where its proteolytic activity is required to maintain the efficiency of photosynthetic machinery during stress. Here, we demonstrate that Deg2 exhibits dual protease-chaperone activities, and we present the hexameric structure of Deg2 complexed with co-purified peptides. The structure shows that Deg2 contains a unique second PDZ domain (PDZ2) following a conventional PDZ domain (PDZ1), with PDZ2 orchestrating the cage assembly of Deg2. We discovered a conserved internal ligand for PDZ2 that mediates hexamer formation and thus locks the protease in the resting state. These findings provide insight into the diverse modes of PDZ domain-mediated regulation of Deg proteases.
- Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Organizational Affiliation: