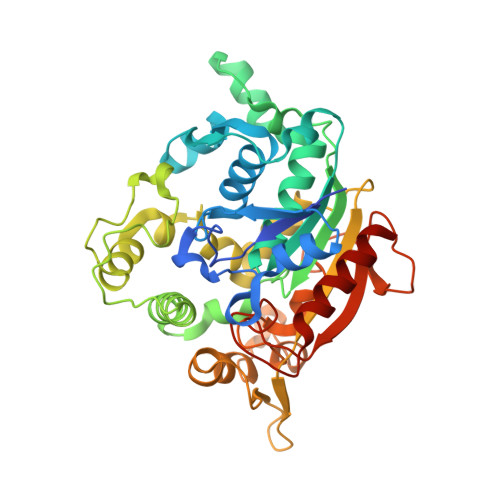
3D Structure Elucidation of Thermostable L2 Lipase from Thermophilic Bacillus sp. L2.
Abd Rahman, R.N., Shariff, F.M., Basri, M., Salleh, A.B.(2012) Int J Mol Sci 13: 9207-9217
- PubMed: 22942761
- DOI: https://doi.org/10.3390/ijms13079207
- Primary Citation of Related Structures:
4FDM - PubMed Abstract:
The crystallization of proteins makes it possible to determine their structure by X-ray crystallography, and is therefore important for the analysis of protein structure-function relationships. L2 lipase was crystallized by using the J-tube counter diffusion method. A crystallization consisting of 20% PEG 6000, 50 mM MES pH 6.5 and 50 mM NaCl was found to be the best condition to produce crystals with good shape and size (0.5 × 0.1 × 0.2 mm). The protein concentration used for the crystallization was 3 mg/mL. L2 lipase crystal has two crystal forms, Shape 1 and Shape 2. Shape 2 L2 lipase crystal was diffracted at 1.5 Å and the crystal belongs to the orthorhombic space group P2(1)2(1)2(1), with unit-cell parameters a = 72.0, b = 81.8, c = 83.4 Å, α = β = γ = 90°. There is one molecule per asymmetric unit and the solvent content of the crystals is 56.9%, with a Matthew's coefficient of 2.85 Å Da(-1). The 3D structure of L2 lipase revealed topological organization of α/β-hydrolase fold consisting of 11 β-strands and 13 α-helices. Ser-113, His-358 and Asp-317 were assigned as catalytic triad residues. One Ca(2+) and one Zn(2+) were found in the L2 lipase molecule.
- Faculty of Biotechnology and Biomolecular Sciences, Enzyme and Microbial Technology Research, Universiti Putra Malaysia, UPM Serdang, Selangor 43400, Malaysia.
Organizational Affiliation: