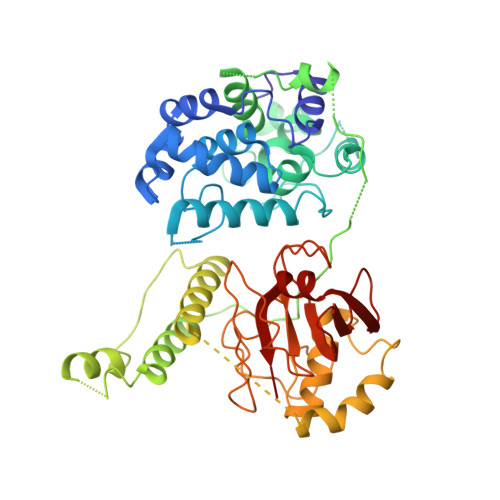
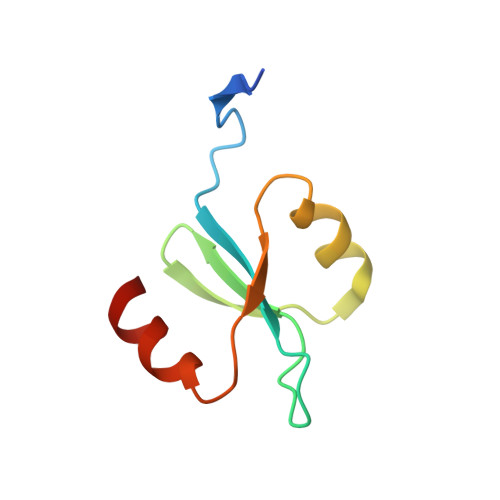
X-ray crystal structure of the streptococcal specific phage lysin PlyC.
McGowan, S., Buckle, A.M., Mitchell, M.S., Hoopes, J.T., Gallagher, D.T., Heselpoth, R.D., Shen, Y., Reboul, C.F., Law, R.H., Fischetti, V.A., Whisstock, J.C., Nelson, D.C.(2012) Proc Natl Acad Sci U S A 109: 12752-12757
- PubMed: 22807482
- DOI: https://doi.org/10.1073/pnas.1208424109
- Primary Citation of Related Structures:
4F87, 4F88 - PubMed Abstract:
Bacteriophages deploy lysins that degrade the bacterial cell wall and facilitate virus egress from the host. When applied exogenously, these enzymes destroy susceptible microbes and, accordingly, have potential as therapeutic agents. The most potent lysin identified to date is PlyC, an enzyme assembled from two components (PlyCA and PlyCB) that is specific for streptococcal species. Here the structure of the PlyC holoenzyme reveals that a single PlyCA moiety is tethered to a ring-shaped assembly of eight PlyCB molecules. Structure-guided mutagenesis reveals that the bacterial cell wall binding is achieved through a cleft on PlyCB. Unexpectedly, our structural data reveal that PlyCA contains a glycoside hydrolase domain in addition to the previously recognized cysteine, histidine-dependent amidohydrolases/peptidases catalytic domain. The presence of eight cell wall-binding domains together with two catalytic domains may explain the extraordinary potency of the PlyC holoenyzme toward target bacteria.
- Department of Biochemistry and Molecular Biology, Monash University, Clayton 3800, Australia.
Organizational Affiliation: