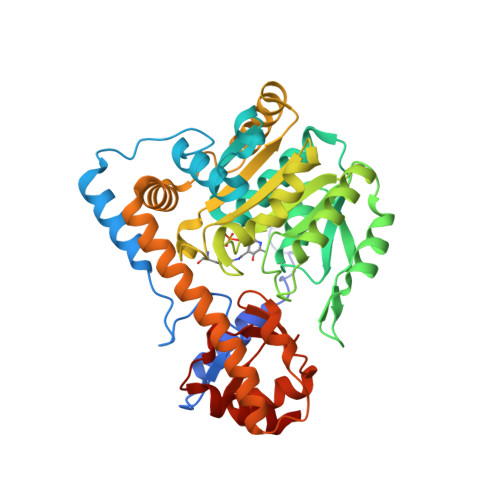
Janus: an algorithm for ranking functional importance of residues from protein sequence alignments.
Addington, T.A., Mertz, R.W., Siegel, J.B., Thompson, J.M., Fisher, A.J., Filkov, V., Fleischman, N., Suen, A., Zhang, C., Toney, M.D.To be published.